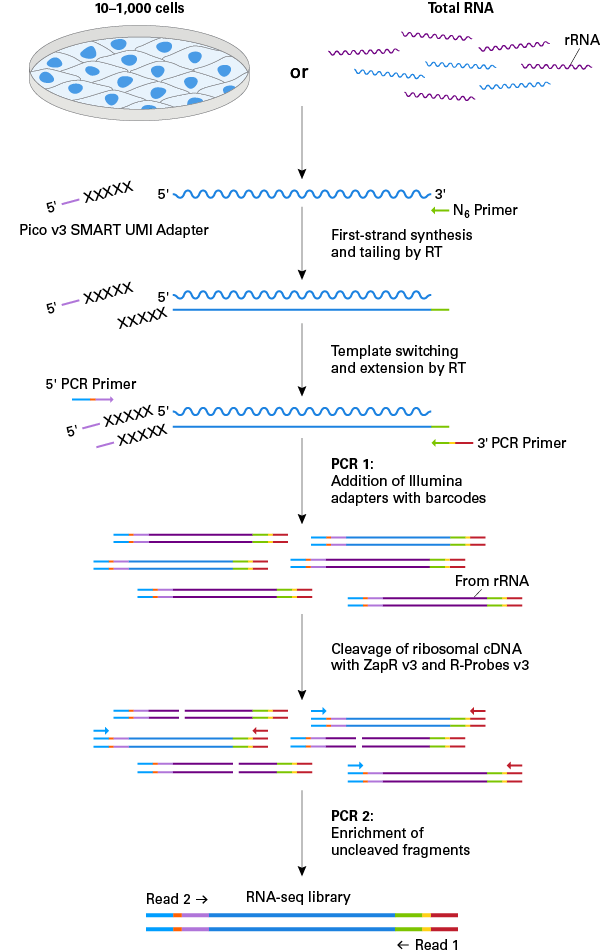
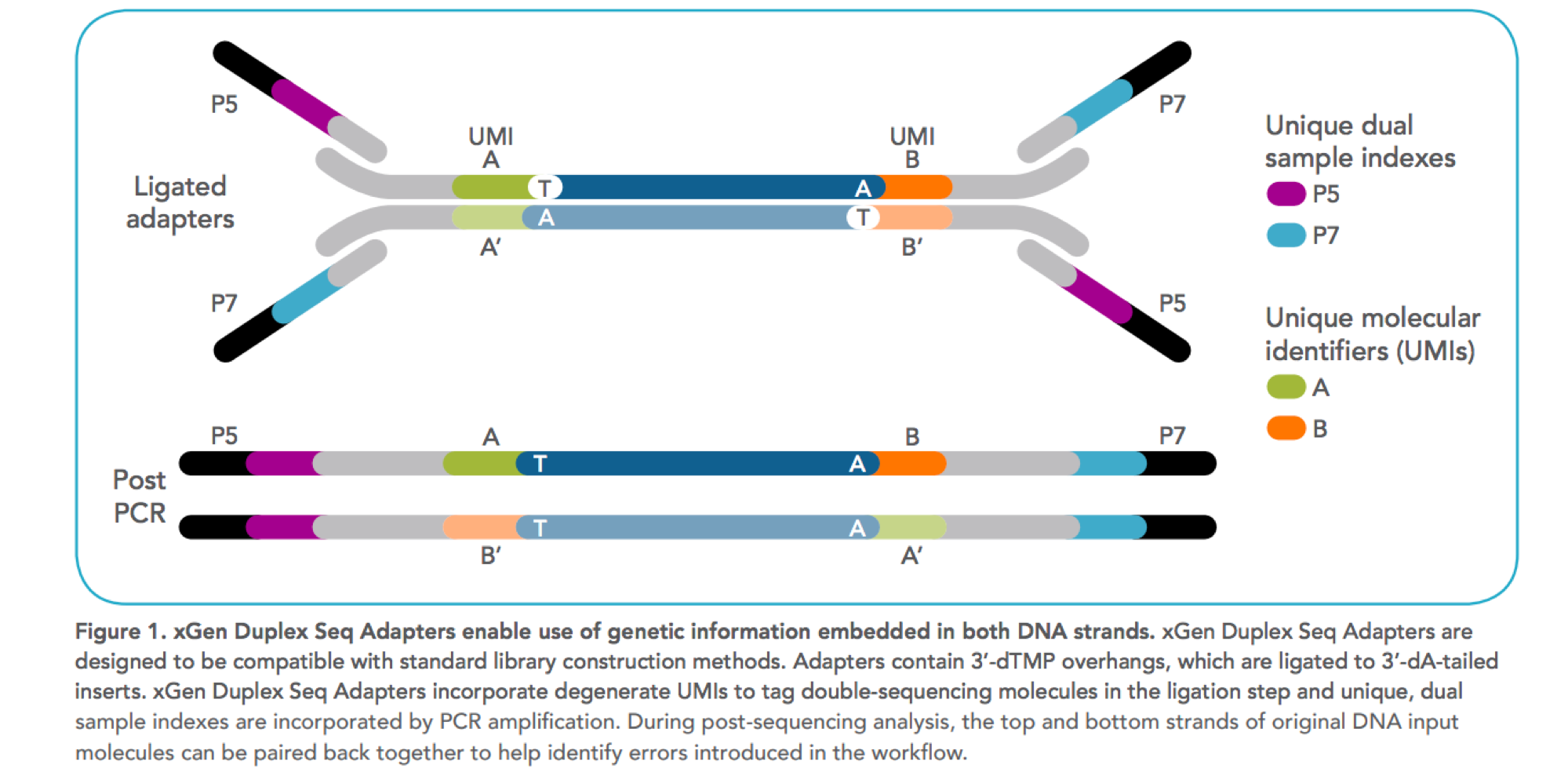
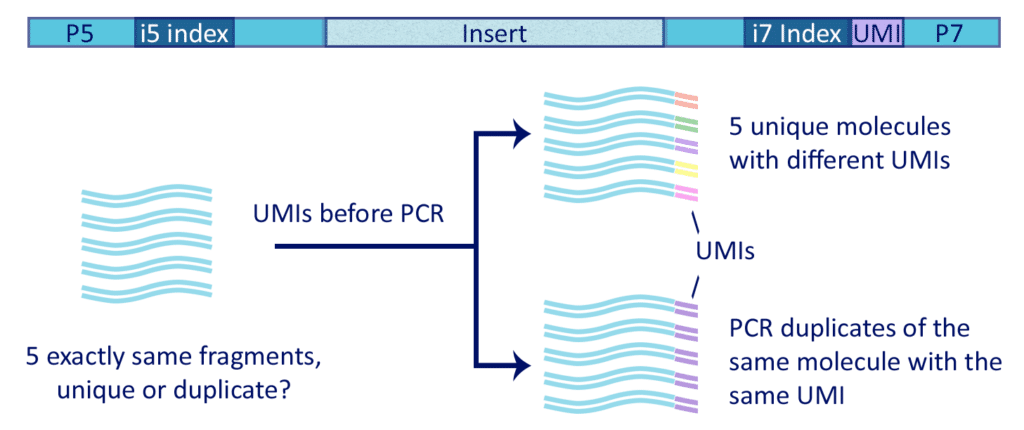
Incorporation of unique molecular identifiers (UMIs) in TruSeq adapters improves the accuracy of quantitative sequencing | RNA-Seq Blog

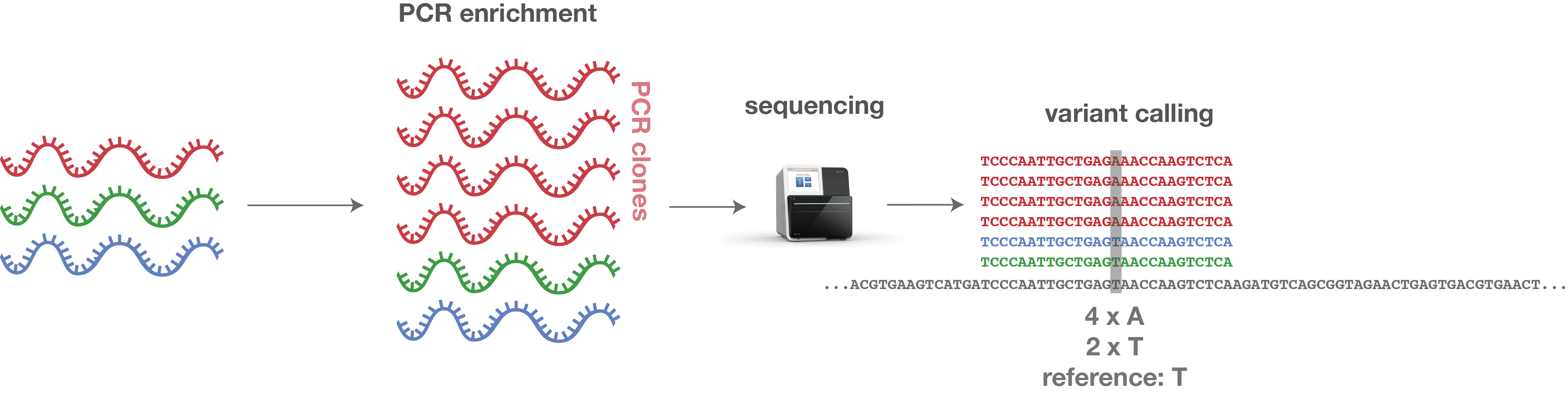
UMI-Gen: a UMI-based reads simulator for variant calling evaluation in paired-end sequencing NGS libraries | bioRxiv
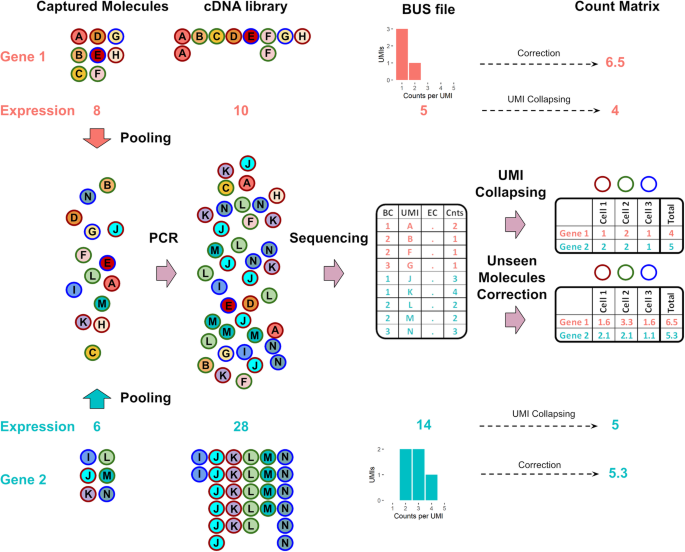
BUTTERFLY: addressing the pooled amplification paradox with unique molecular identifiers in single-cell RNA-seq | Genome Biology | Full Text

IsoSeek – unbiased and UMI-informed sequencing of cell-free miRNAs at single-nucleotide resolution | RNA-Seq Blog
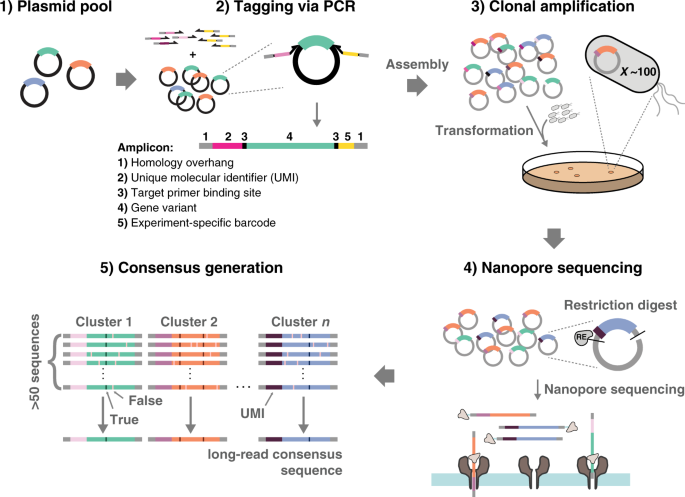
UMI-linked consensus sequencing enables phylogenetic analysis of directed evolution | Nature Communications
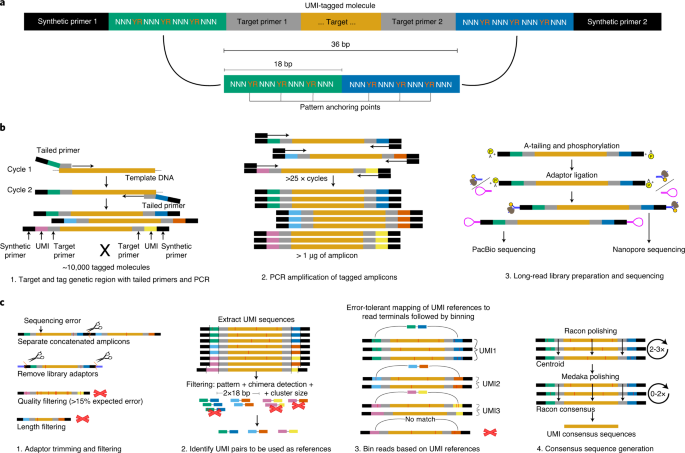
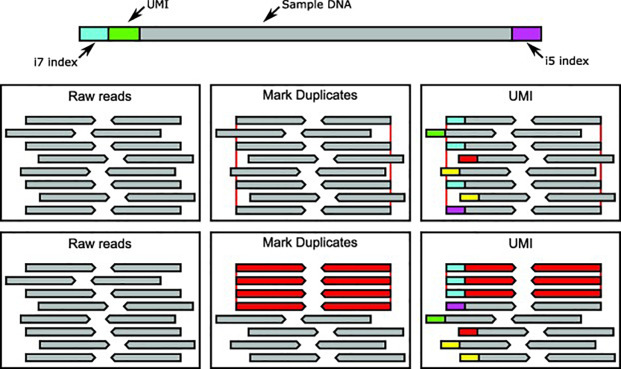
High-accuracy long-read amplicon sequences using unique molecular identifiers with Nanopore or PacBio sequencing | Nature Methods

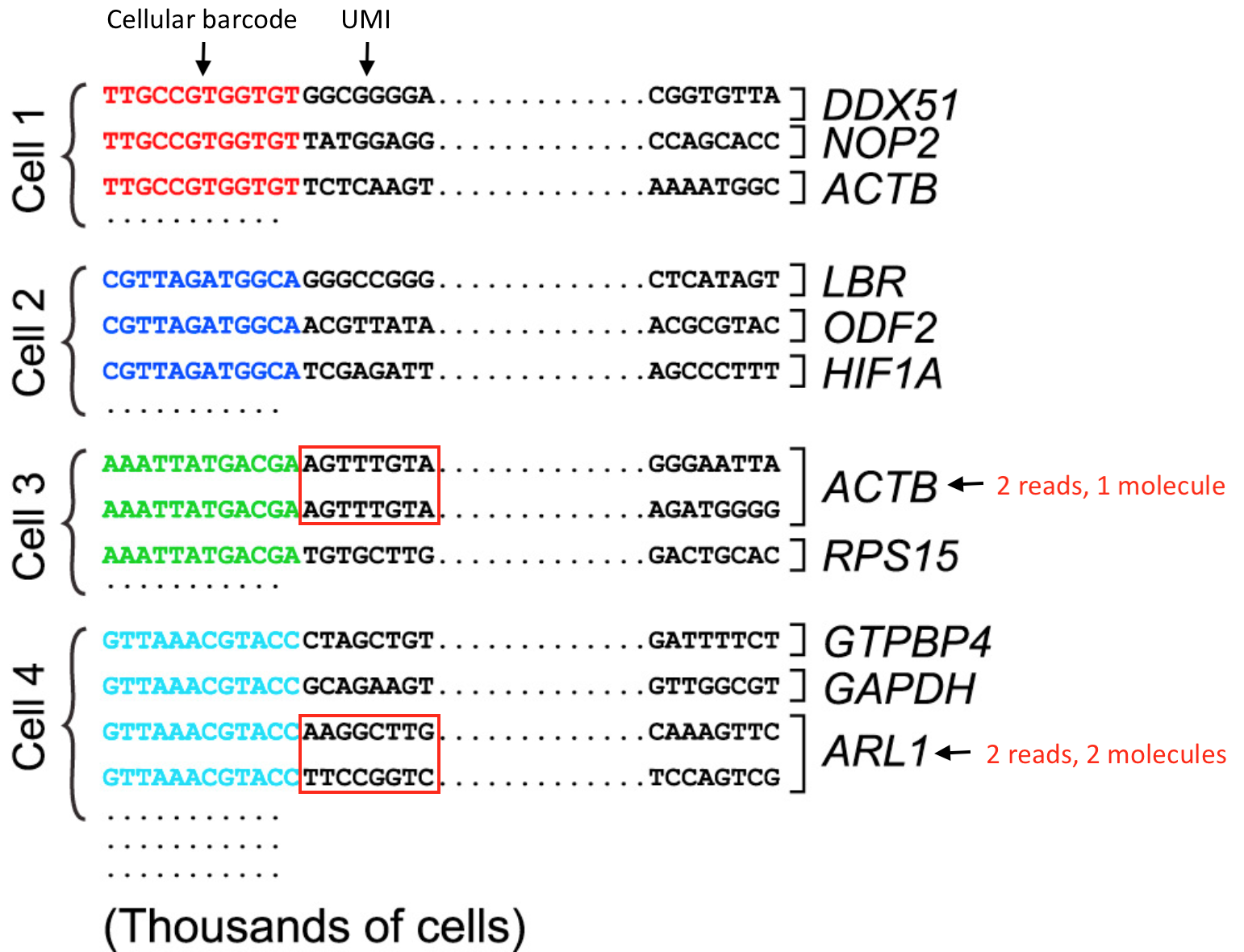
Use of unique molecular identifiers (UMIs). Each strand is an mRNA or... | Download Scientific Diagram