Oxford Nanopore on Twitter: "Cas9 targeted sequencing with Oxford #Nanopore is completely PCR-free, enabling enrichment of very large regions, including low-complexity and GC-rich sequences. Check out the full infographic for more info:
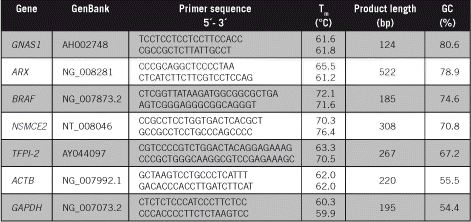
PCR amplification of GC-rich DNA regions using the nucleotide analog N4-methyl-2′-deoxycytidine 5′-triphosphate | BioTechniques

The SSV‐Seq 2.0 PCR‐Free Method Improves the Sequencing of Adeno‐Associated Viral Vector Genomes Containing GC‐Rich Regions and Homopolymers - Lecomte - 2021 - Biotechnology Journal - Wiley Online Library
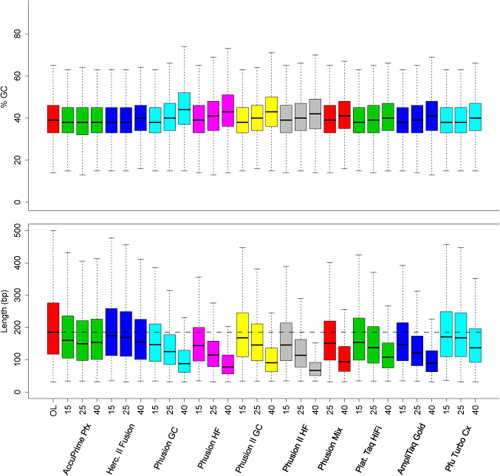
Length and GC-biases during sequencing library amplification: A comparison of various polymerase-buffer systems with ancient and modern DNA sequencing libraries | BioTechniques