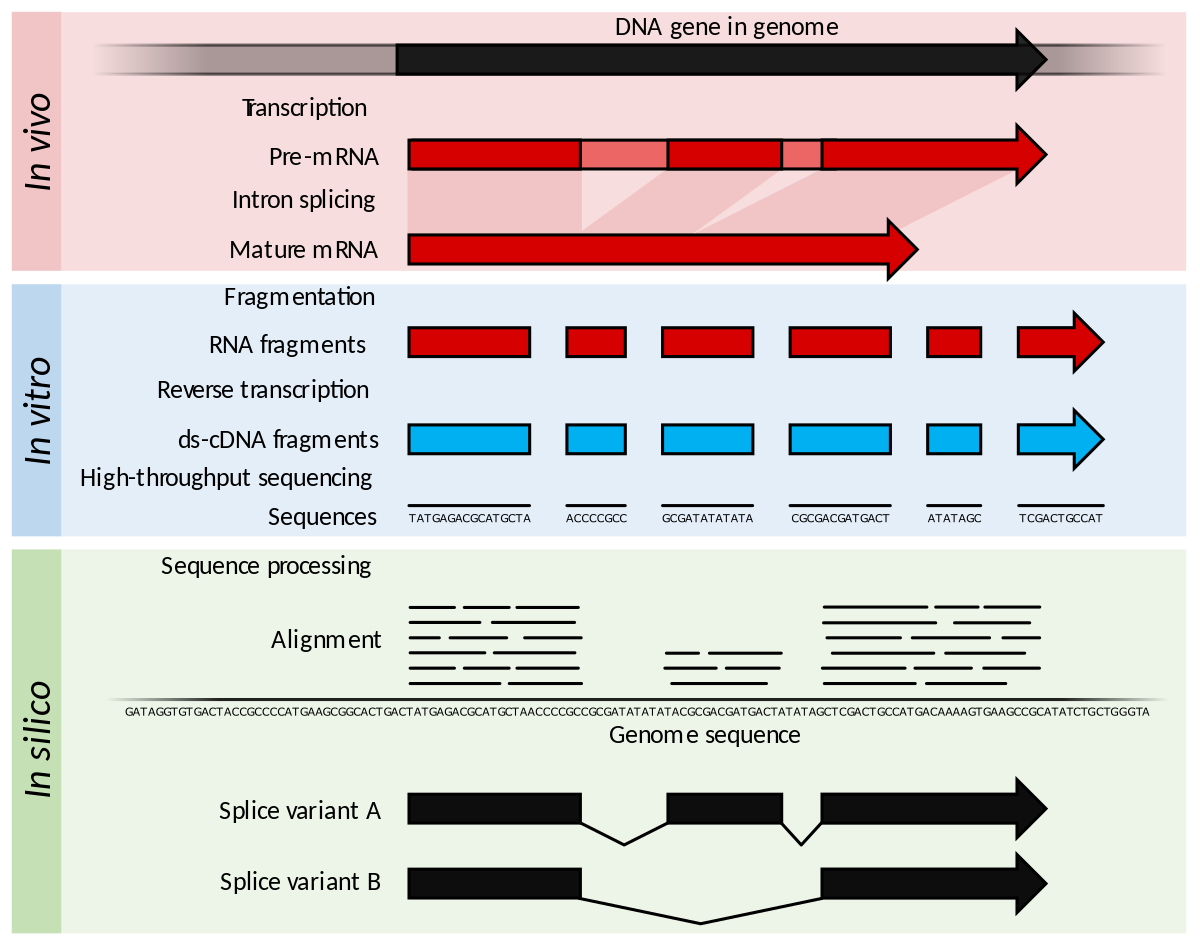
Difference between sequencing Coverage and depth. Depth vs Coverage. Why they are important? - YouTube

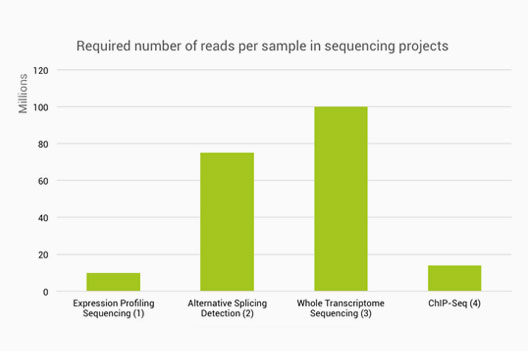
Determination of the number of reads needed for each RNA-Seq protocol... | Download Scientific Diagram

Read coverage of genome sequencing and RNA-seq data. a Read coverage of... | Download Scientific Diagram
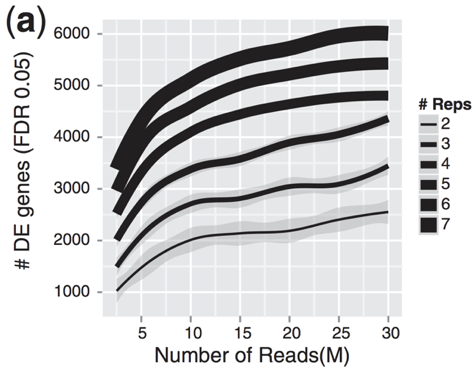
Experimental design considerations | Introduction to RNA-Seq using high-performance computing - ARCHIVED

The trade-off between increased multiplexing and decreasing sequencing depth in smallRNA-Seq | RNA-Seq Blog
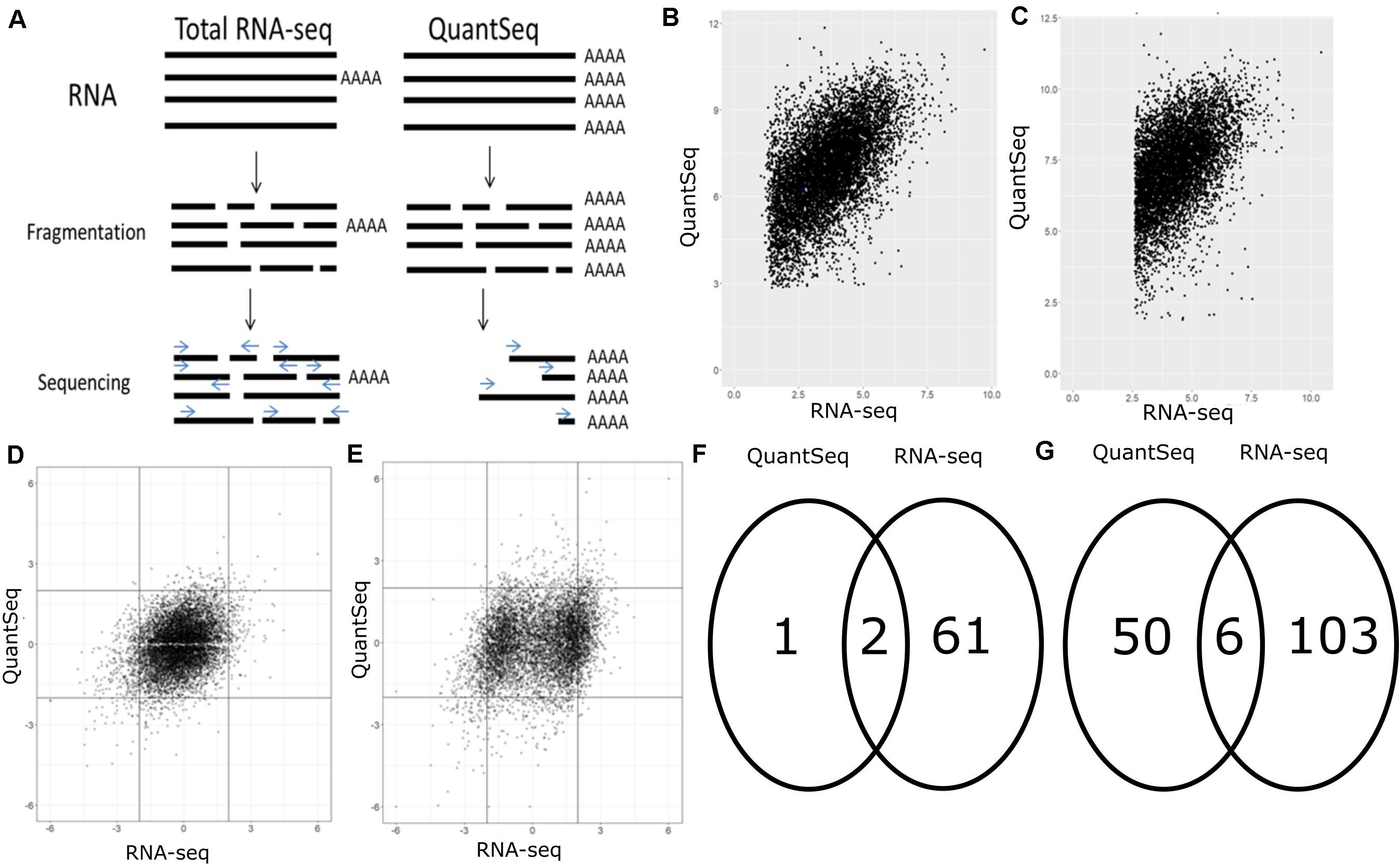
Frontiers | A Comparison of Low Read Depth QuantSeq 3′ Sequencing to Total RNA-Seq in FUS Mutant Mice