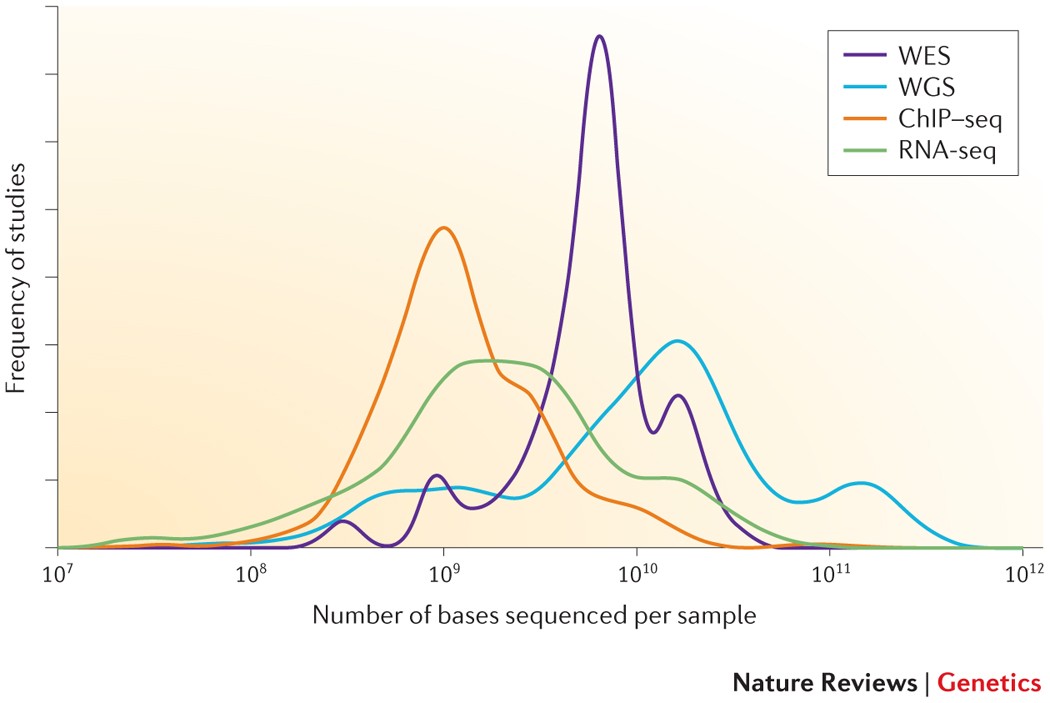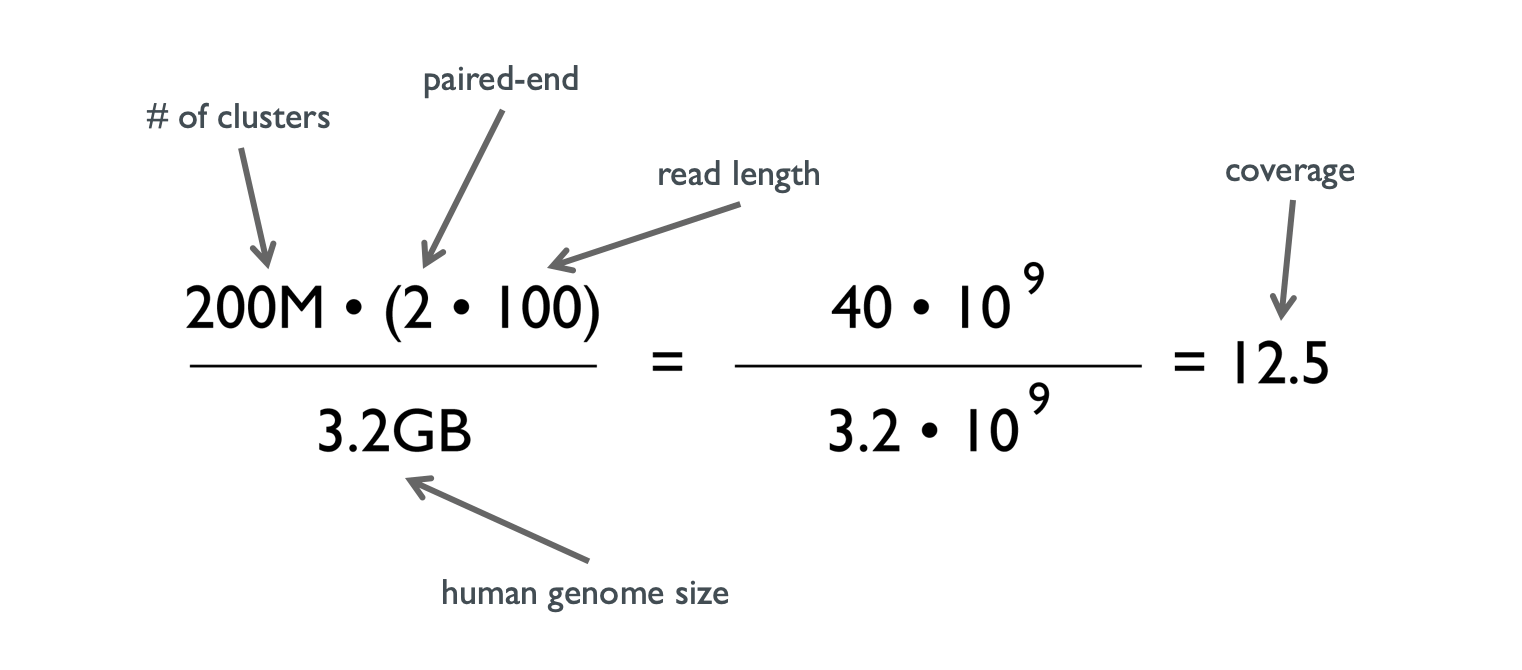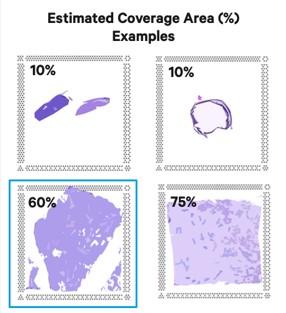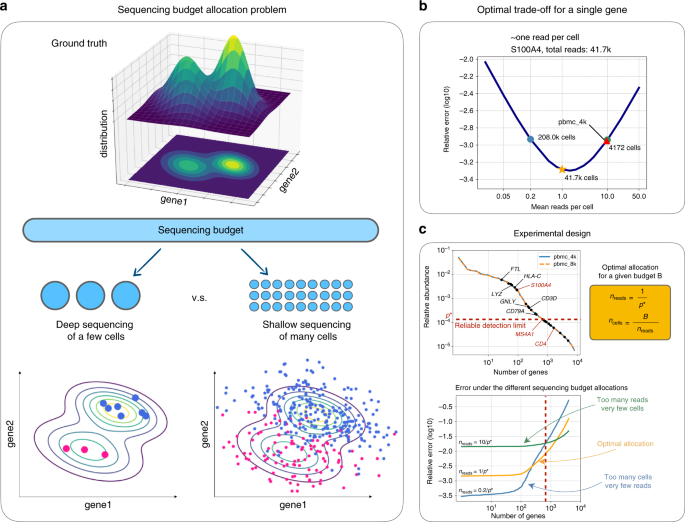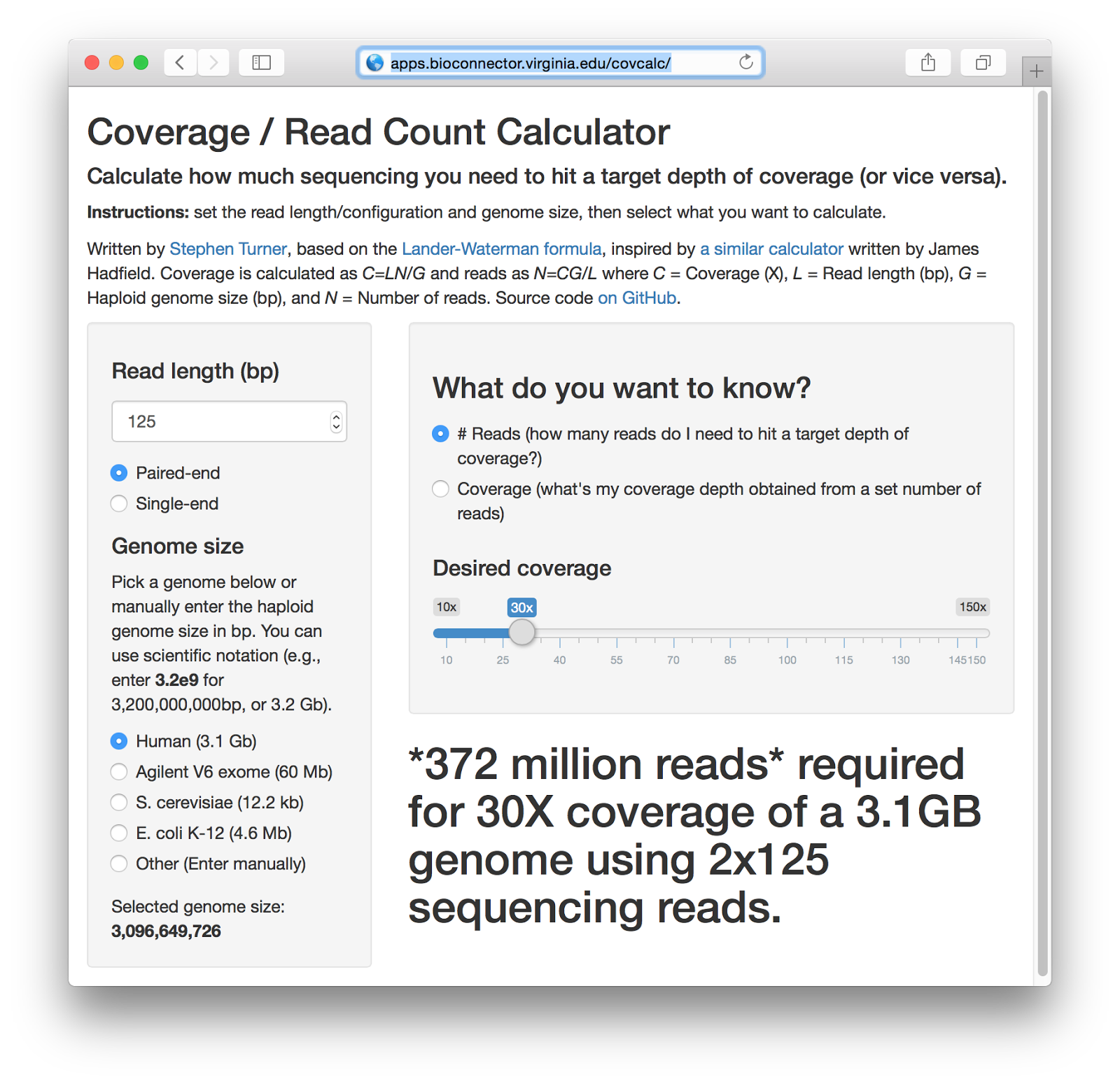
Getting Genetics Done: Covcalc: Shiny App for Calculating Coverage Depth or Read Counts for Sequencing Experiments
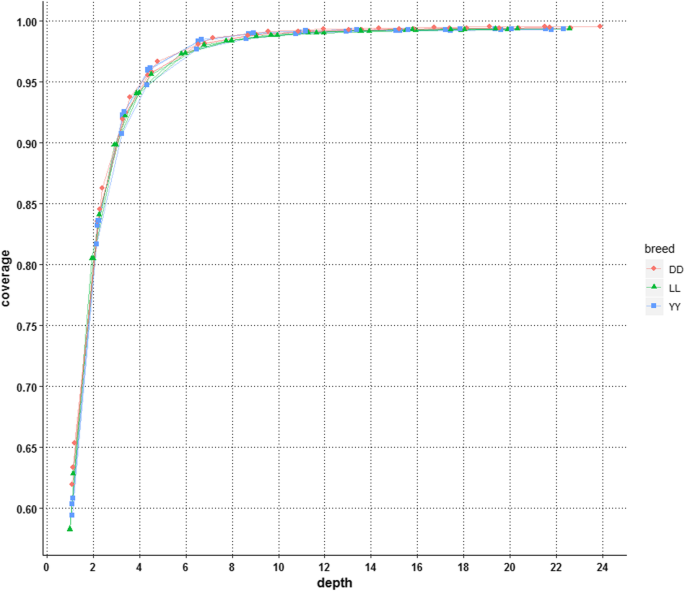
Optimal sequencing depth design for whole genome re-sequencing in pigs | BMC Bioinformatics | Full Text
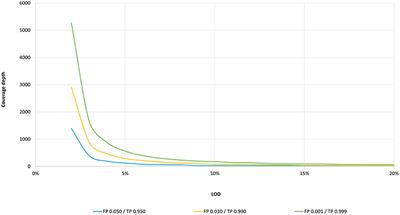
Frontiers | Standardization of Sequencing Coverage Depth in NGS: Recommendation for Detection of Clonal and Subclonal Mutations in Cancer Diagnostics

Covcalc: Shiny App for Calculating Coverage Depth or Read Counts for Sequencing Experiments | R-bloggers

A beginner's guide to low‐coverage whole genome sequencing for population genomics - Lou - 2021 - Molecular Ecology - Wiley Online Library
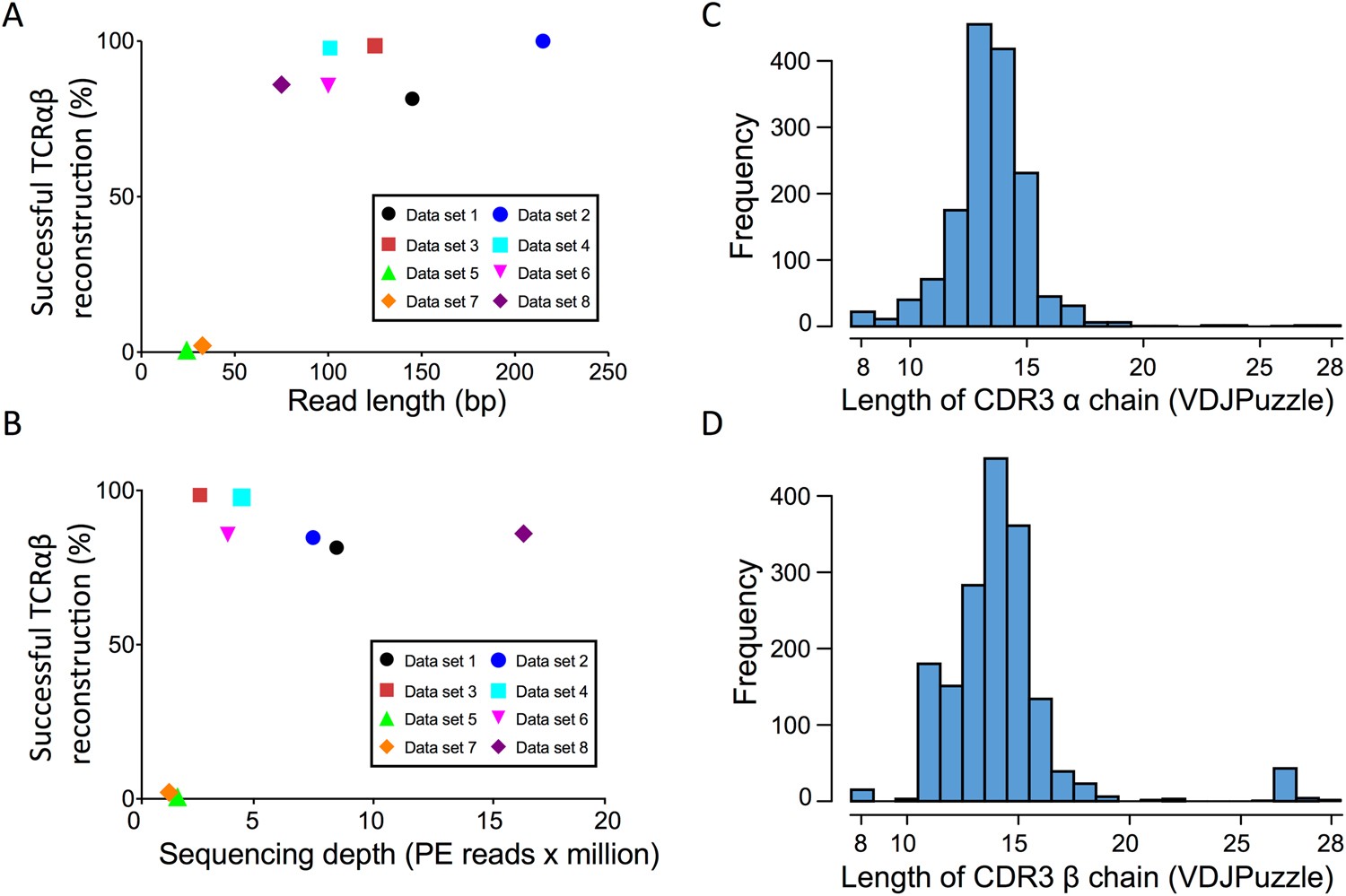
Impact of sequencing depth and read length on single cell RNA sequencing data of T cells | Scientific Reports