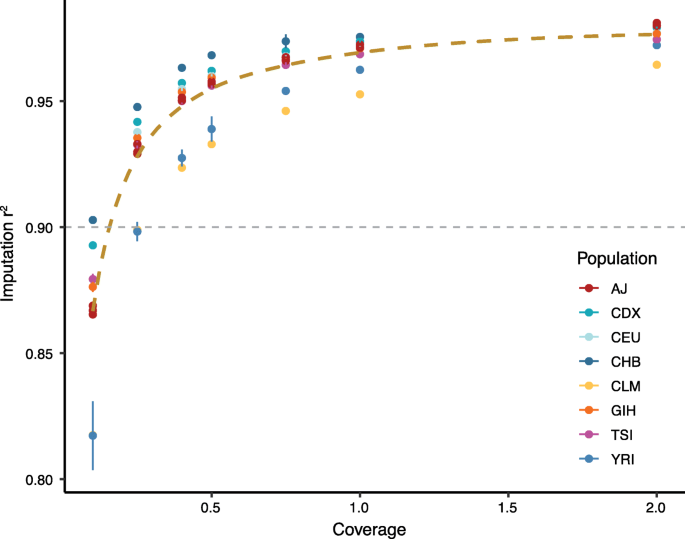
Low coverage whole genome sequencing enables accurate assessment of common variants and calculation of genome-wide polygenic scores | Genome Medicine | Full Text
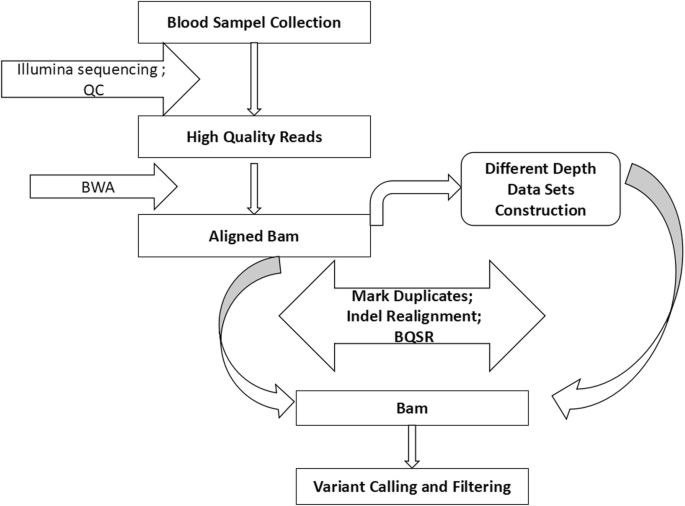
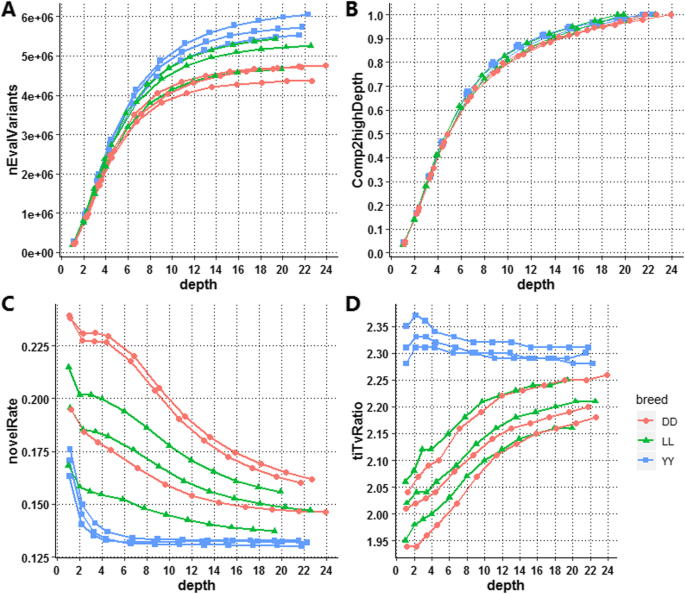
Optimal sequencing depth design for whole genome re-sequencing in pigs | BMC Bioinformatics | Full Text
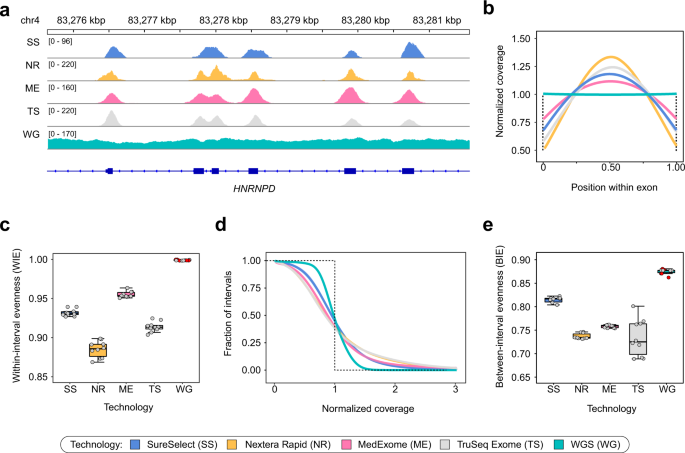
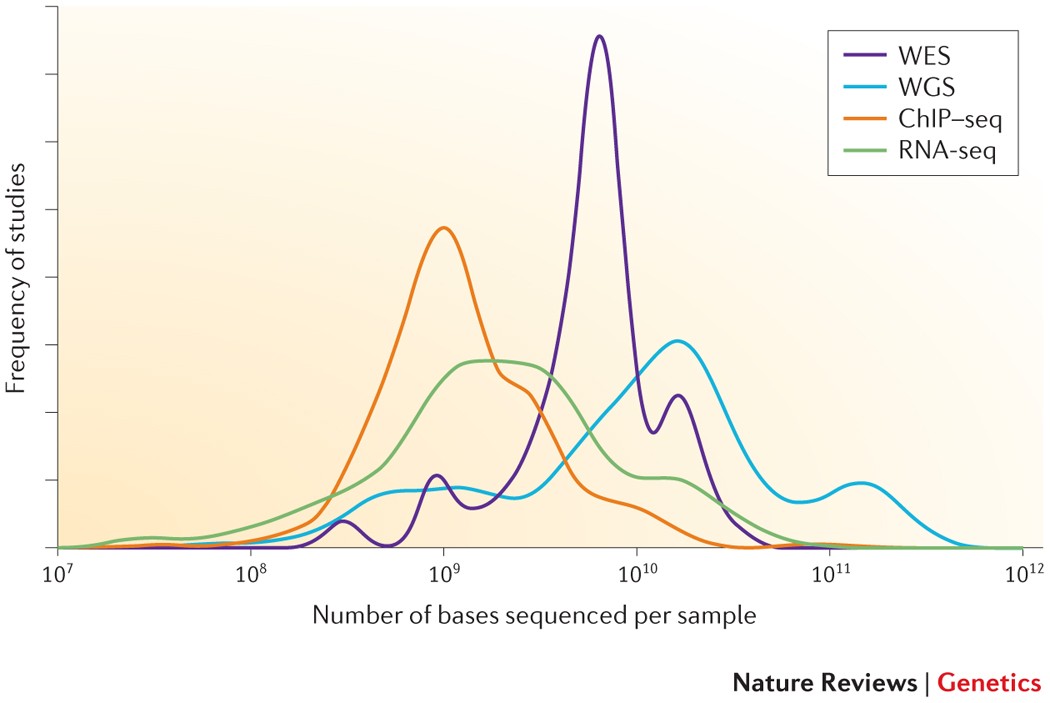
Systematic dissection of biases in whole-exome and whole-genome sequencing reveals major determinants of coding sequence coverage | Scientific Reports
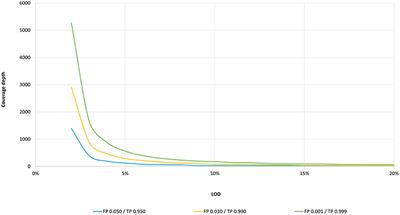
Frontiers | Standardization of Sequencing Coverage Depth in NGS: Recommendation for Detection of Clonal and Subclonal Mutations in Cancer Diagnostics

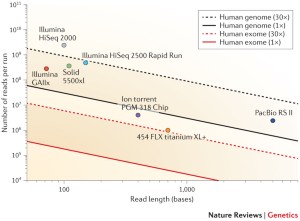
Mean mapped depth and coverage of diagnostic genomic regions according... | Download Scientific Diagram

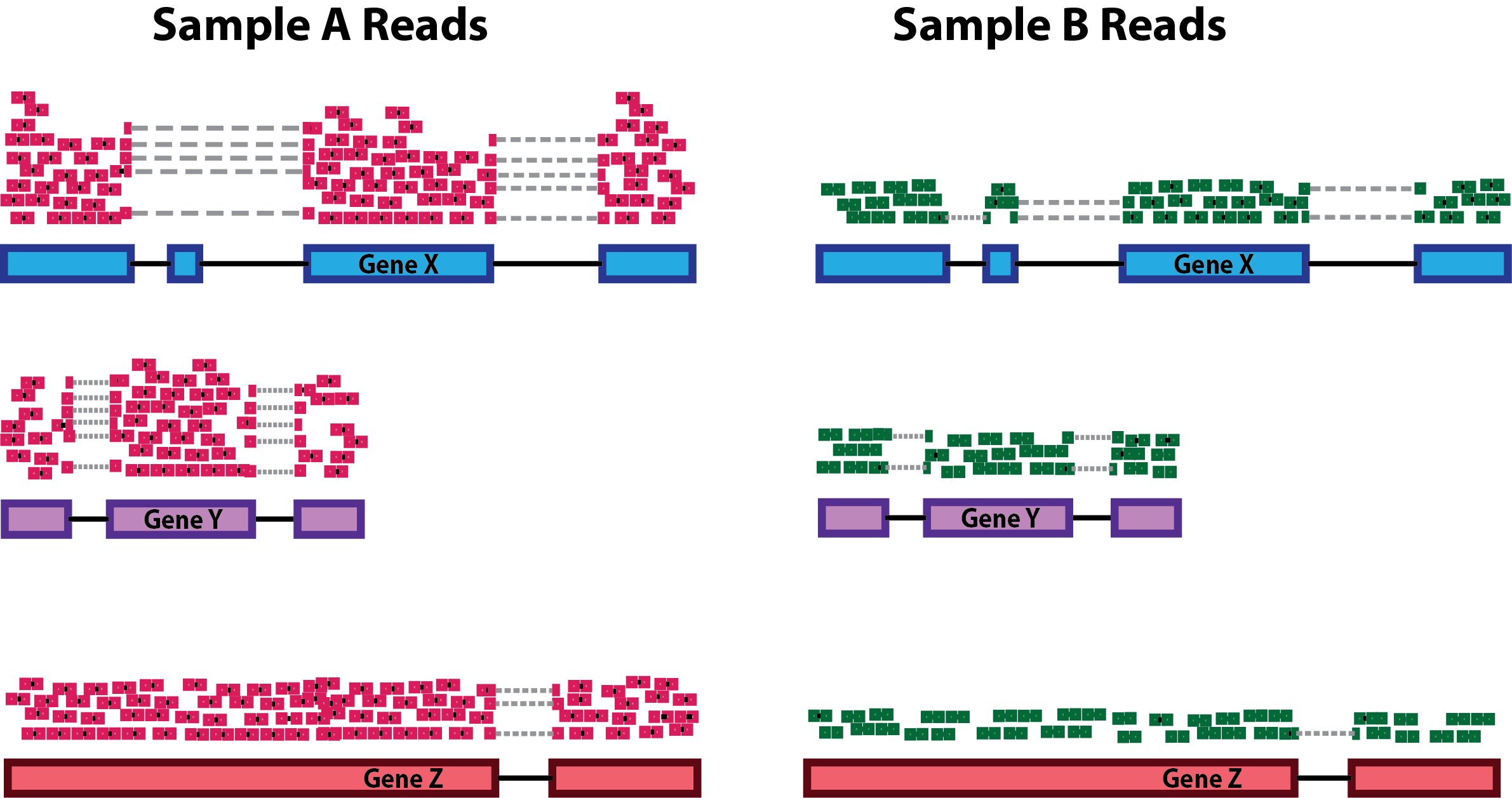
Detecting, Categorizing, and Correcting Coverage Anomalies of RNA-Seq Quantification - ScienceDirect

Systematic dissection of biases in whole-exome and whole-genome sequencing reveals major determinants of coding sequence coverage | Scientific Reports