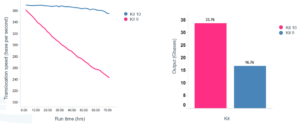
Oxford Nanopore on Twitter: "Introducing the new Sequencing Auxiliary Vials expansion pack; allowing you to split library across multiple flow cells, add library back following a wash, or add additional library to

Minimal requirements for ISO15189 validation and accreditation of three next generation sequencing procedures for SARS-CoV-2 surveillance in clinical setting | Scientific Reports

Water | Free Full-Text | MinION Nanopore Sequencing Accelerates Progress towards Ubiquitous Genetics in Water Research

chemistry-technical-document-CHTD 500 v1 revAA 07jul2016-Any | PDF | Dna Sequencing | Complementary Dna

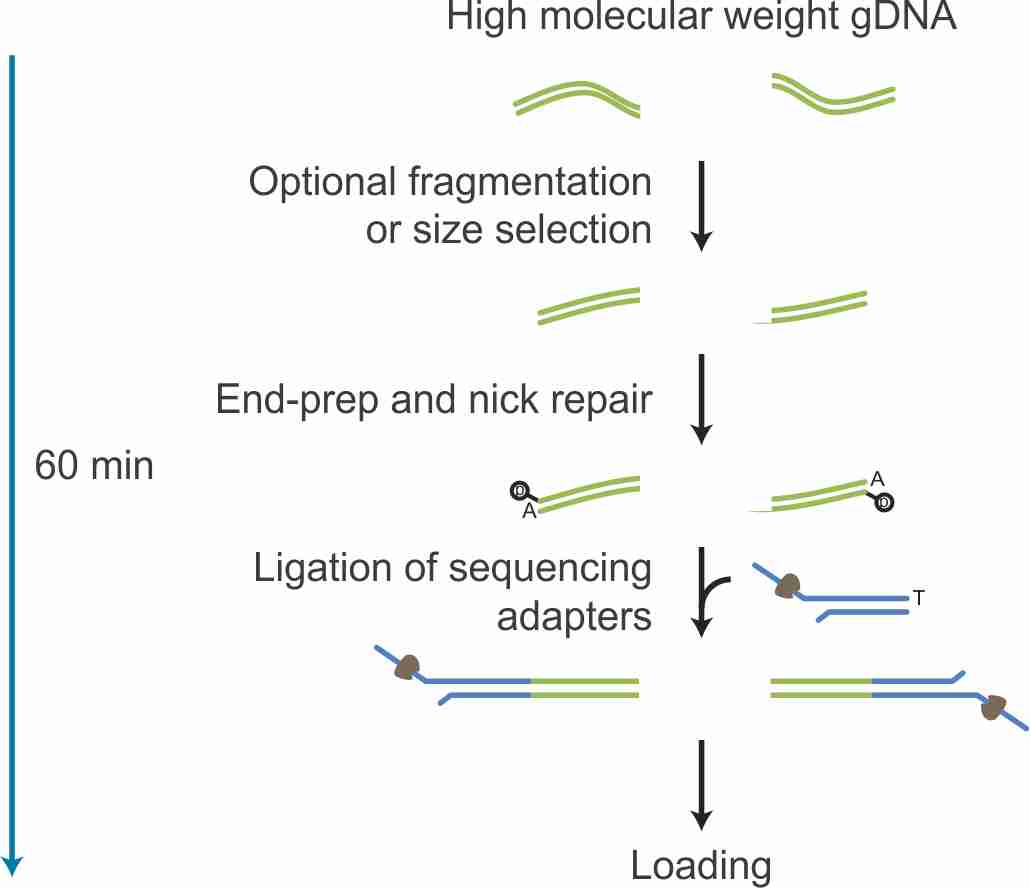
pcr-barcoding-genomic-dna-sqk-lsk110-PBGE12_9111_v110_revI_10Nov2020-minion | PDF | Dna Ligase | Dna Sequencing
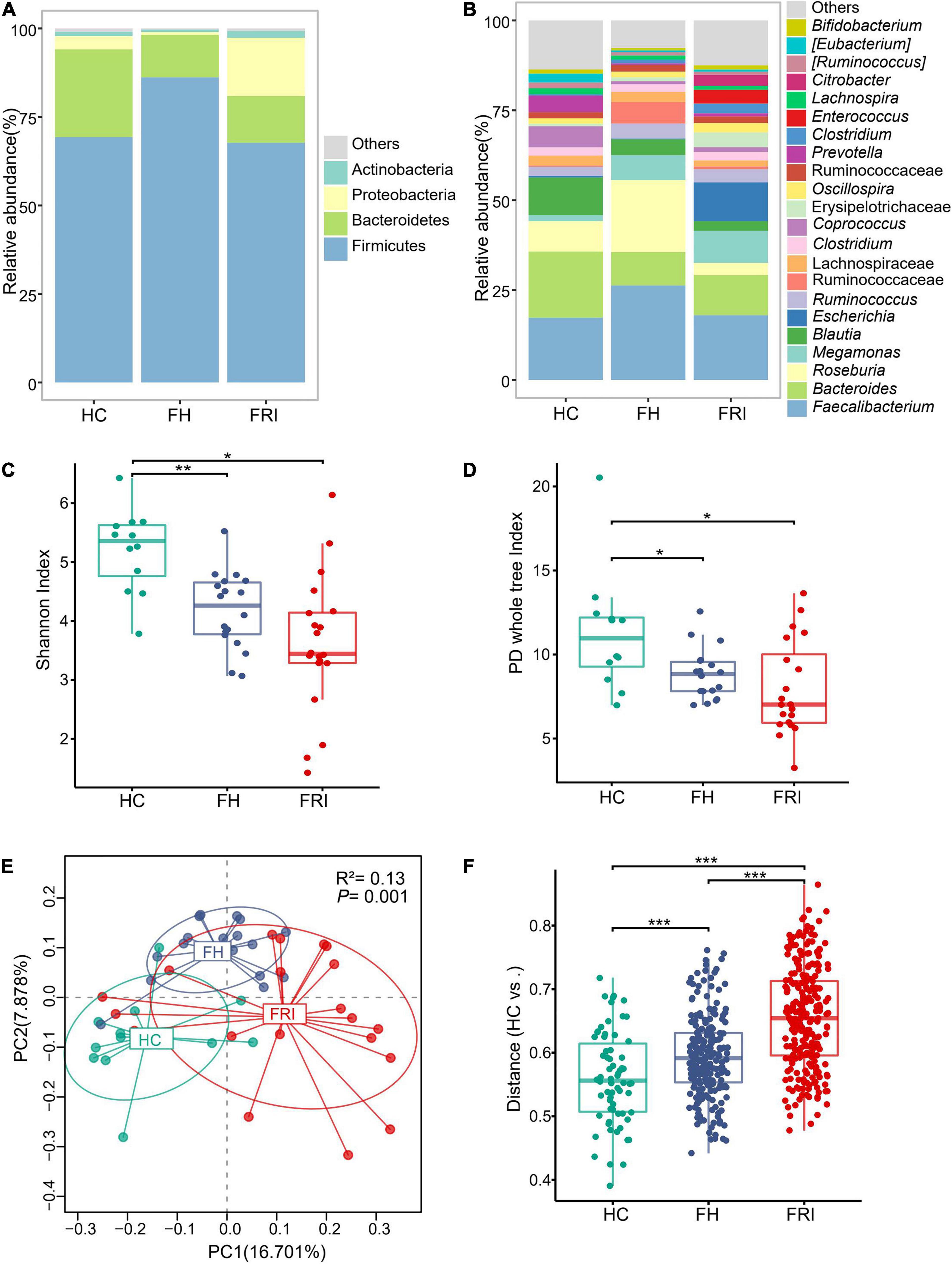
Frontiers | Altered Gut Microbiota as an Auxiliary Diagnostic Indicator for Patients With Fracture-Related Infection