Table 3 from CLIP-seq analysis of multi-mapped reads discovers novel functional RNA regulatory sites in the human transcriptome | Semantic Scholar

Experimental design and data analysis of Ago‐RIP‐Seq experiments for the identification of microRNA targets | Semantic Scholar

Dynamic transcriptomic m5C and its regulatory role in RNA processing - Chen - 2021 - WIREs RNA - Wiley Online Library

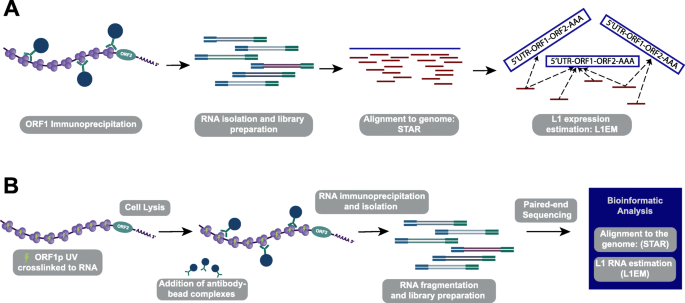
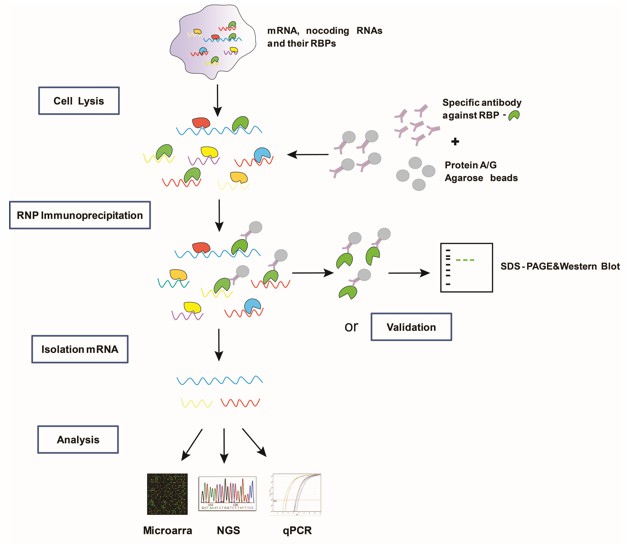
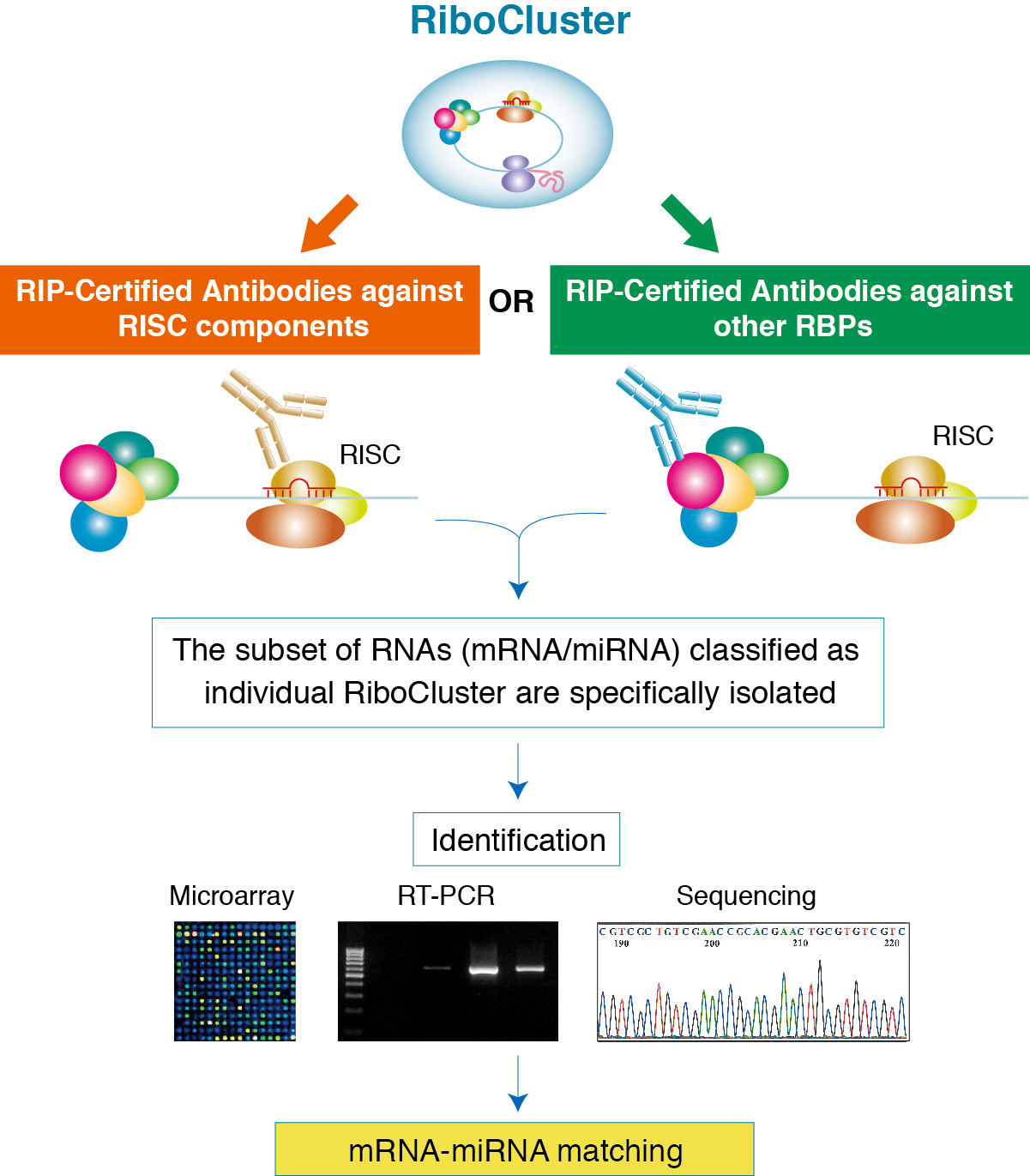
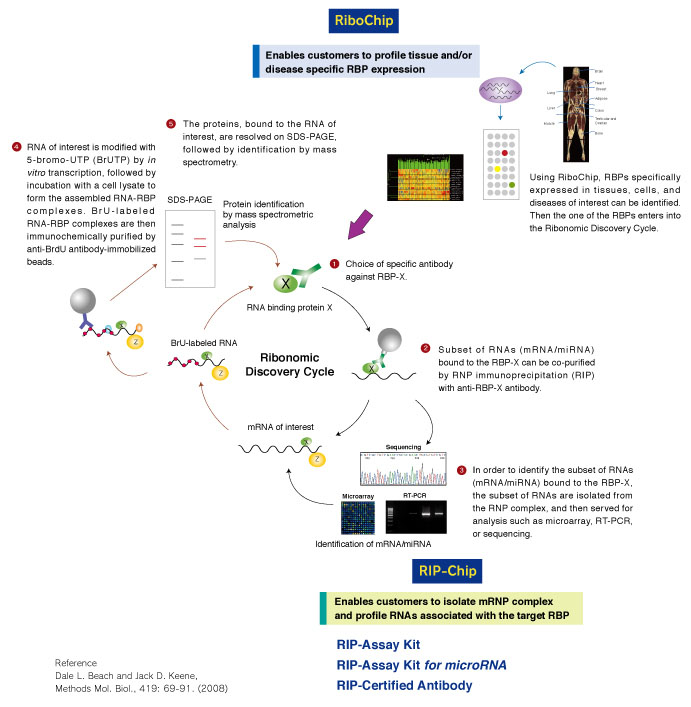
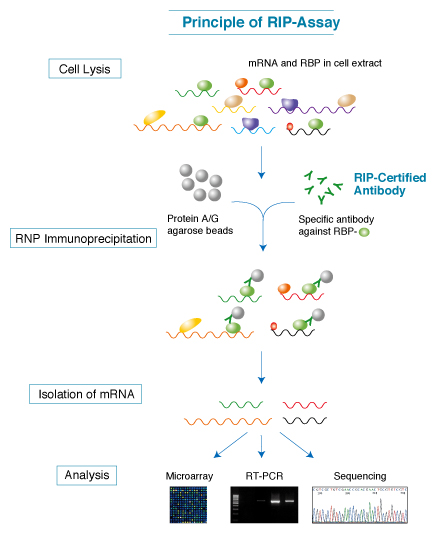
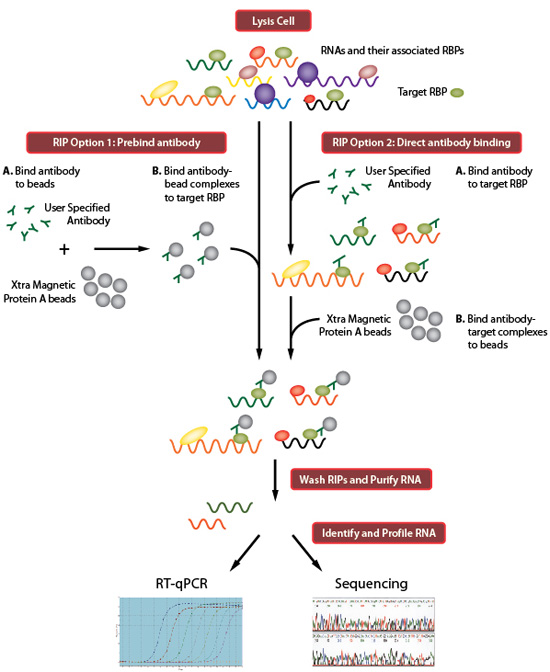
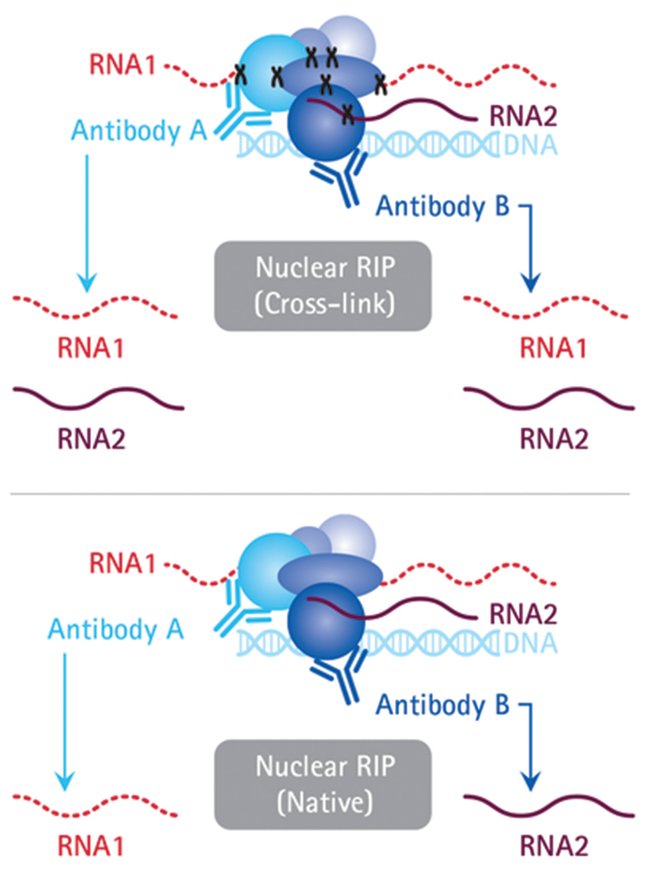
Computational approaches for the analysis of RNA–protein interactions: A primer for biologists - ScienceDirect

Identification of RNA Binding Partners of CRISPR-Cas Proteins in Prokaryotes Using RIP-Seq | SpringerLink

Computational approaches for the analysis of RNA–protein interactions: A primer for biologists - ScienceDirect