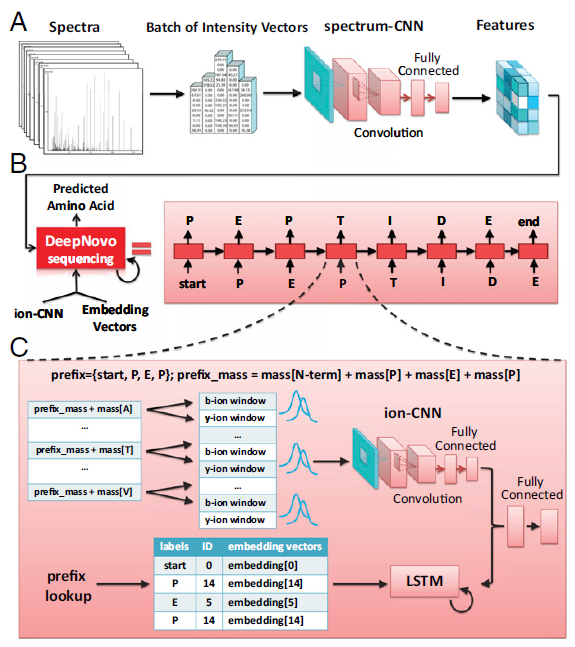
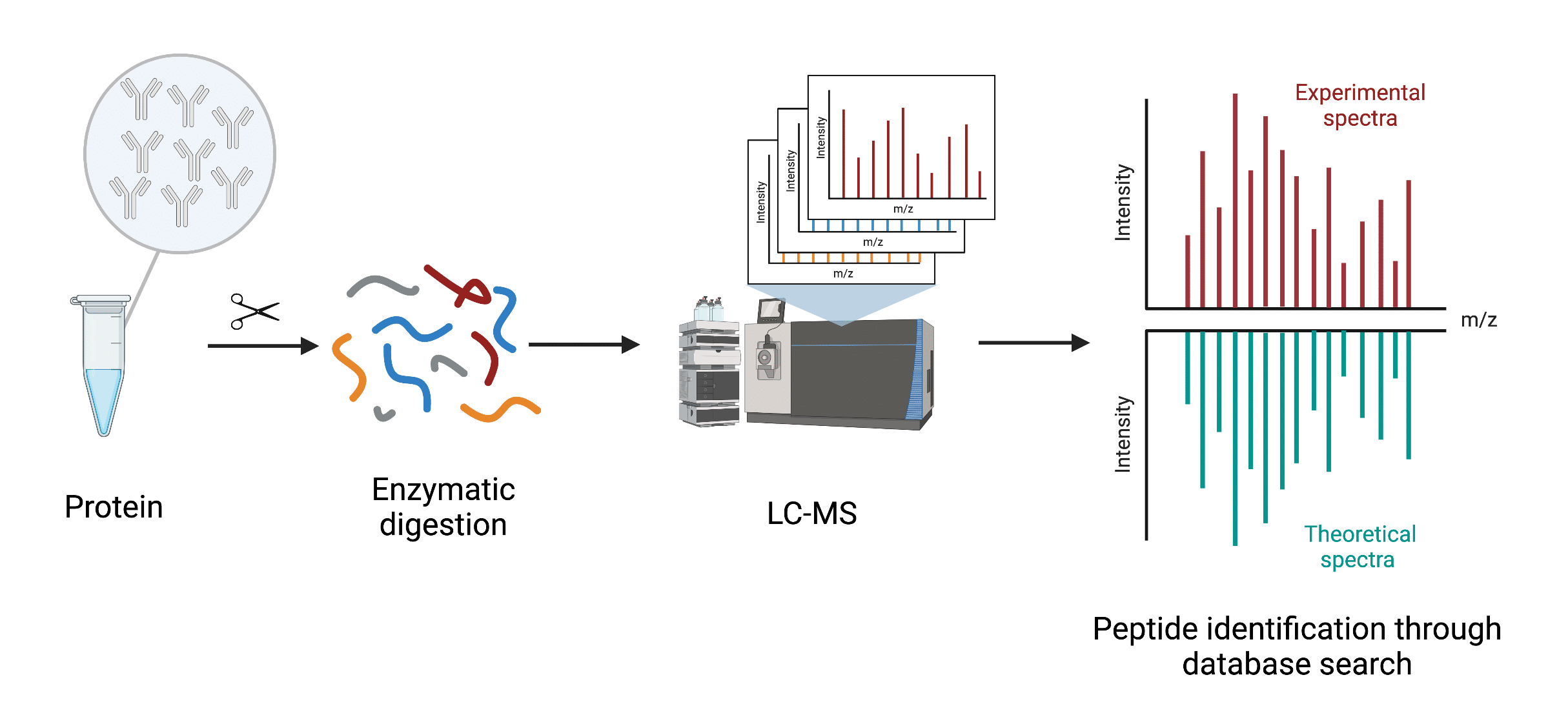
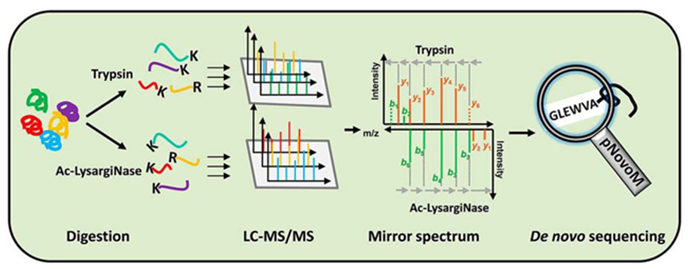
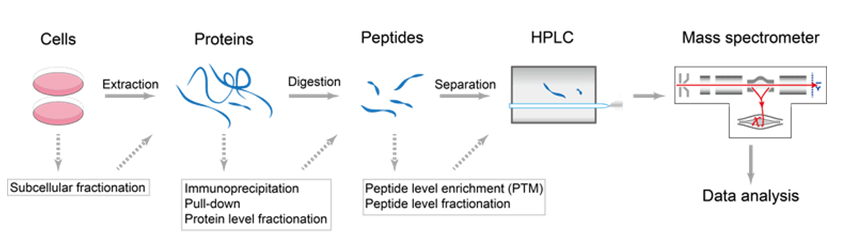
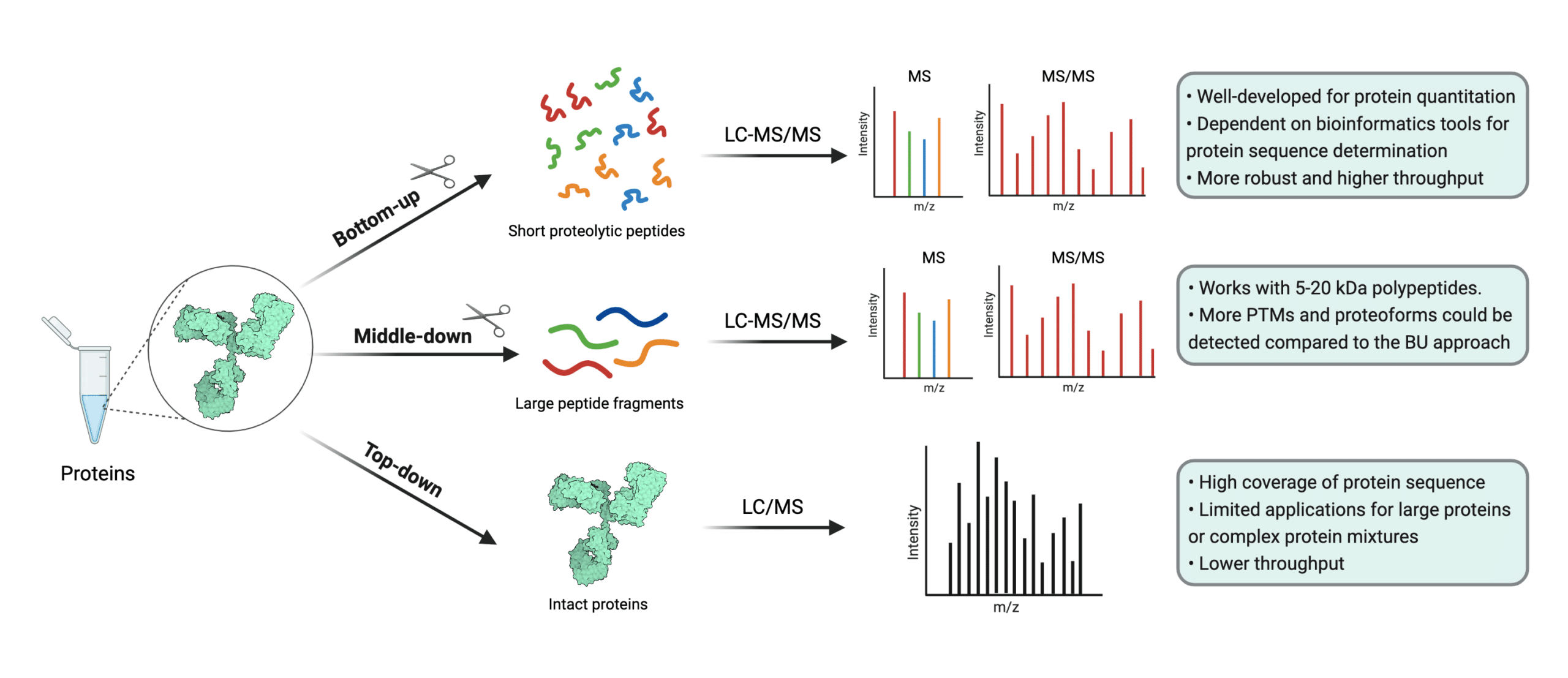
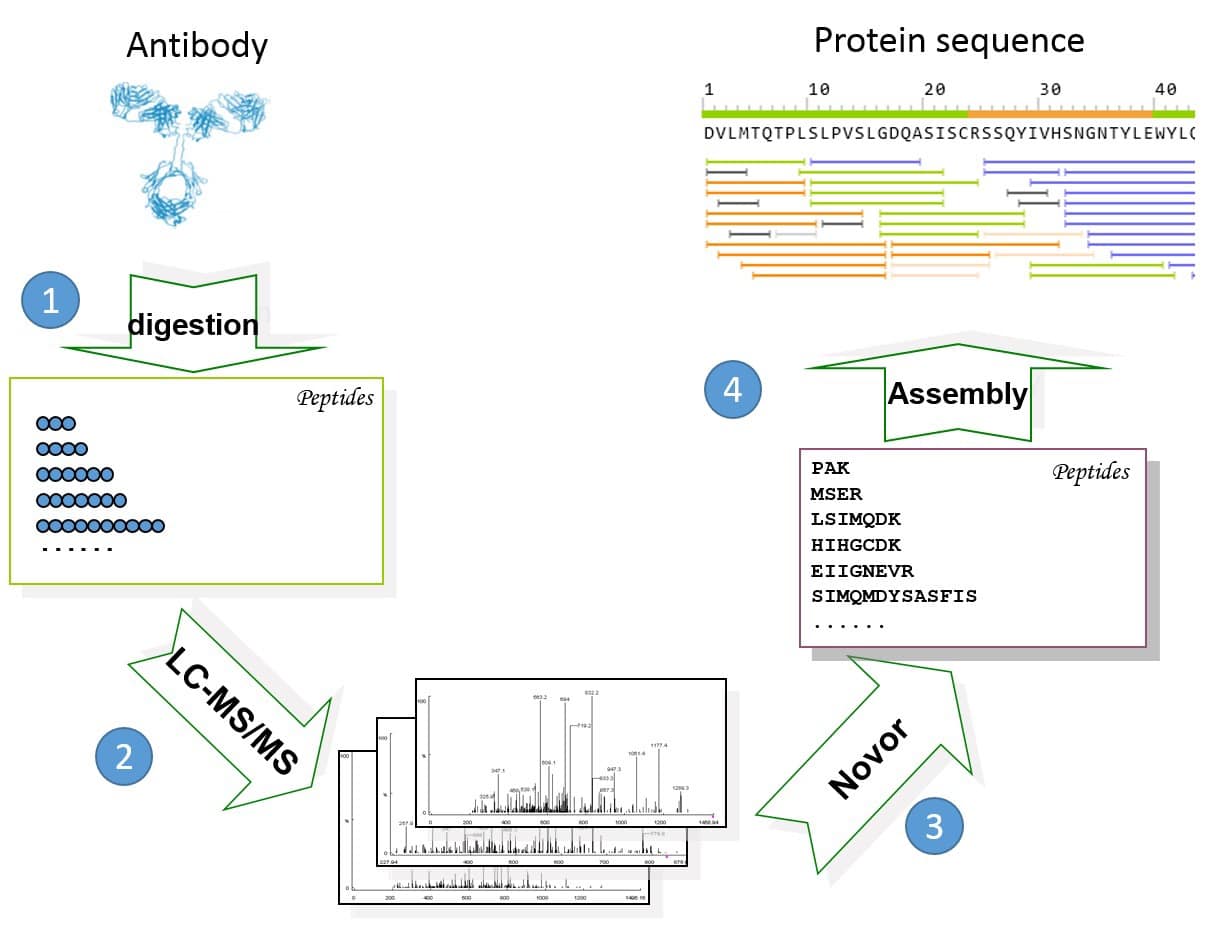
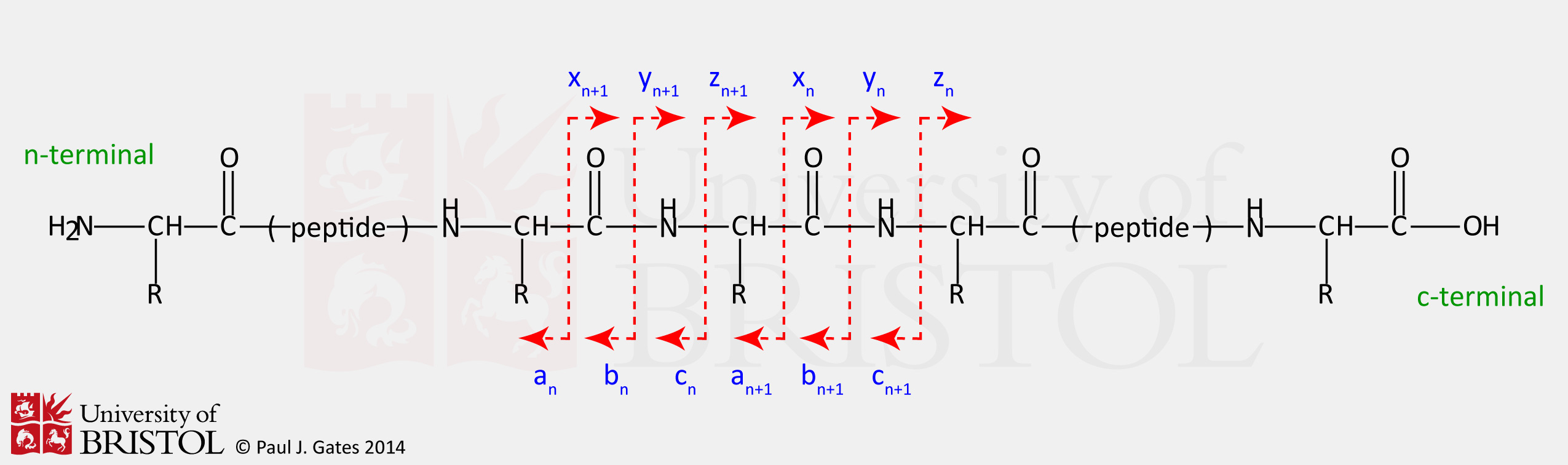
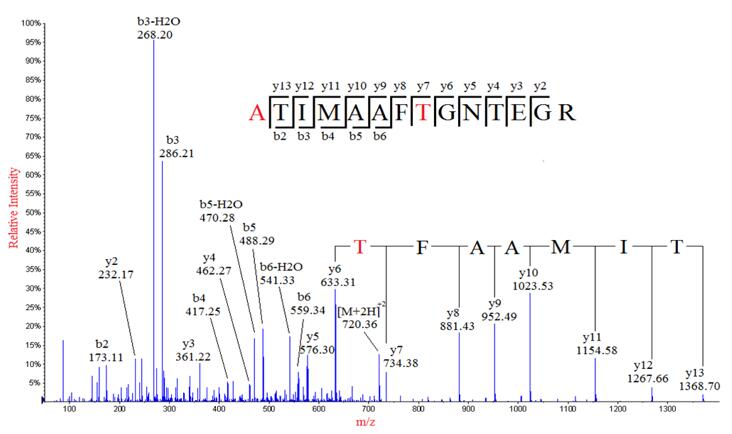
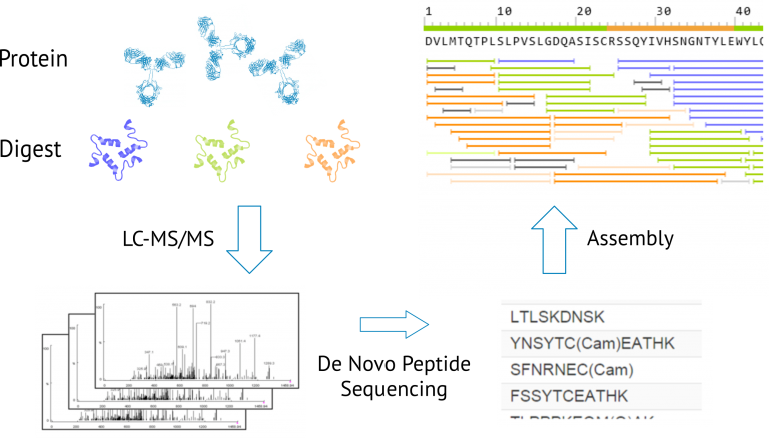
Uncovering Thousands of New Peptides with Sequence-Mask-Search Hybrid De Novo Peptide Sequencing Framework - ScienceDirect

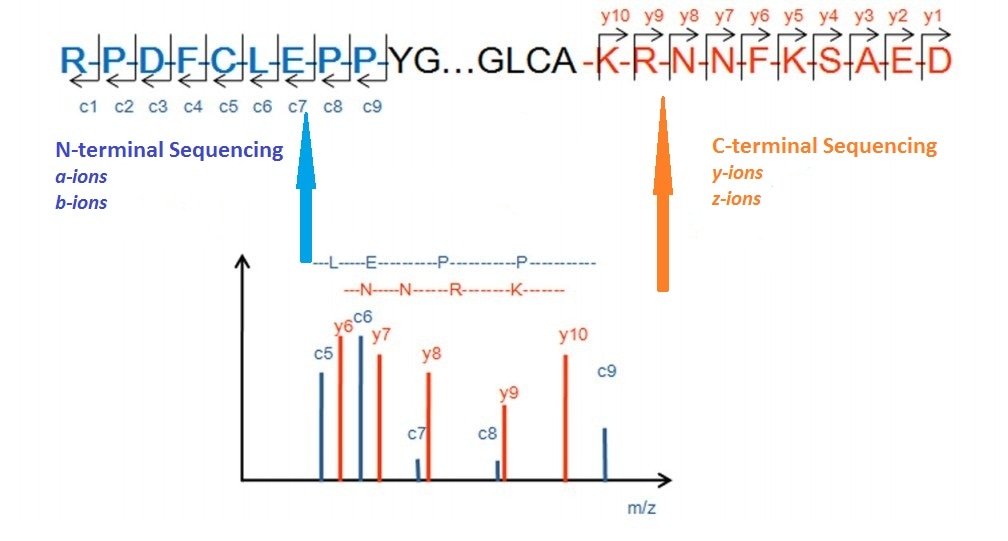
Identification and Sequencing of N-Terminal Peptides in Proteins by LC-Fluorescence-MS/MS: An Approach to Replacement of the Edman Degradation | Analytical Chemistry
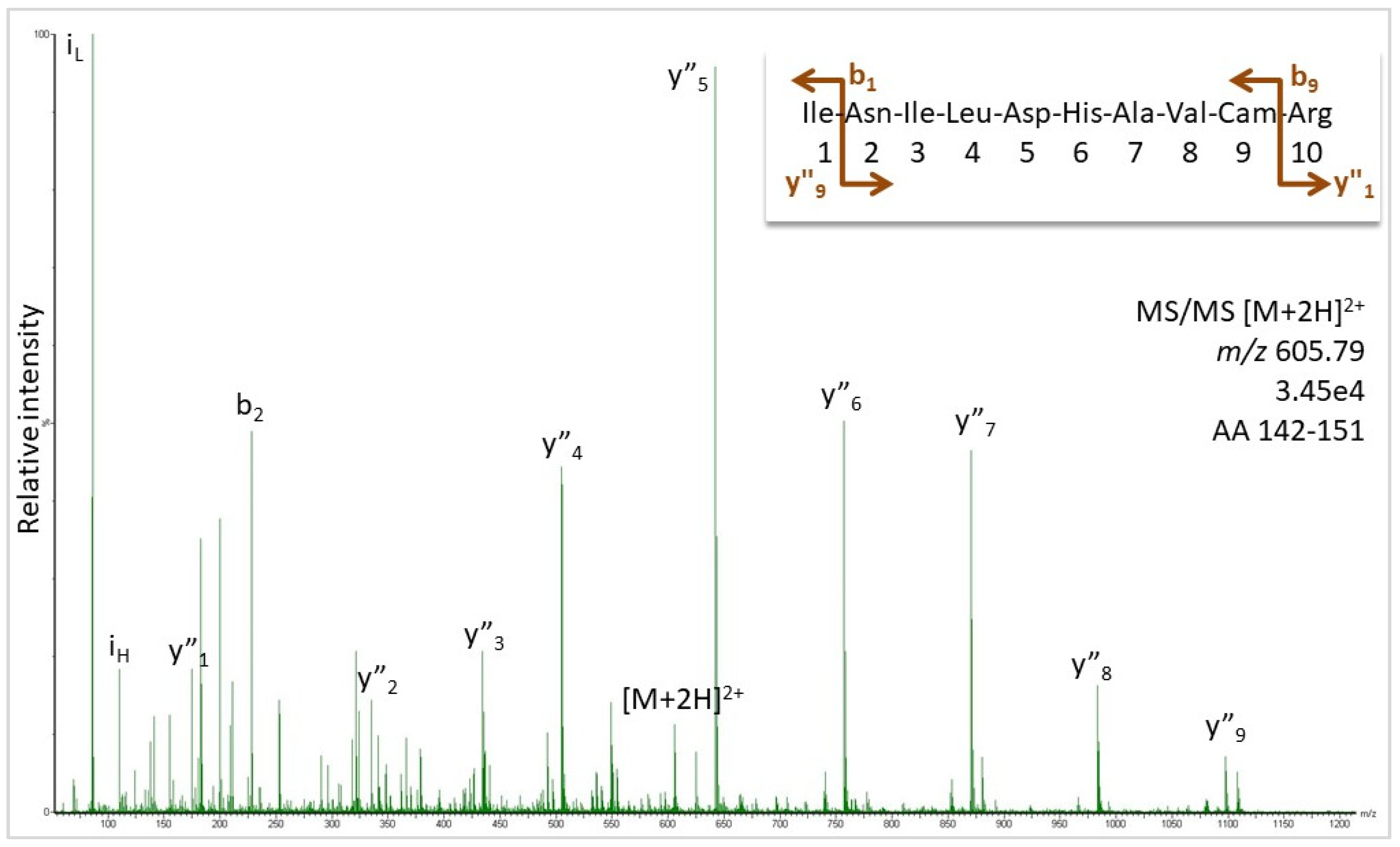
Molecules | Free Full-Text | The Current State-of-the-Art Identification of Unknown Proteins Using Mass Spectrometry Exemplified on De Novo Sequencing of a Venom Protease from Bothrops moojeni

Identification and Sequencing of N-Terminal Peptides in Proteins by LC-Fluorescence-MS/MS: An Approach to Replacement of the Edman Degradation | Analytical Chemistry

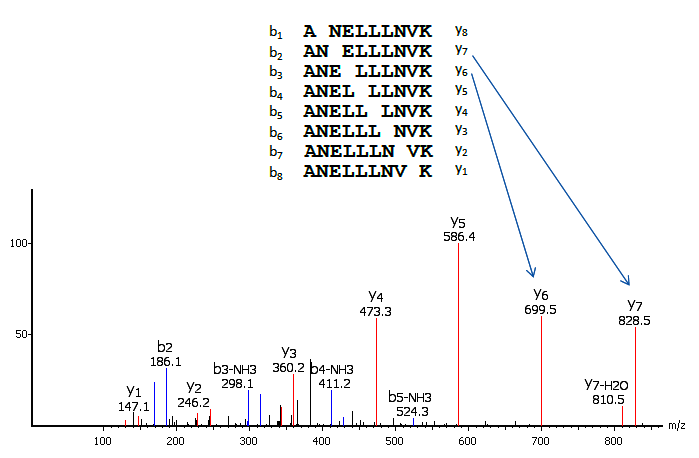
MS/MS for peptide sequencing and identification of oxidative cysteine... | Download Scientific Diagram

Free Radical–Initiated Peptide Sequencing Mass Spectrometry for Phosphopeptide Post-translational Modification Analysis | SpringerLink