Sequence analysis of FUS-NLS. (A). Domain structure of FUS with the... | Download Scientific Diagram

A bipartite NLS sequence on Ci is necessary and sufficient to direct... | Download Scientific Diagram
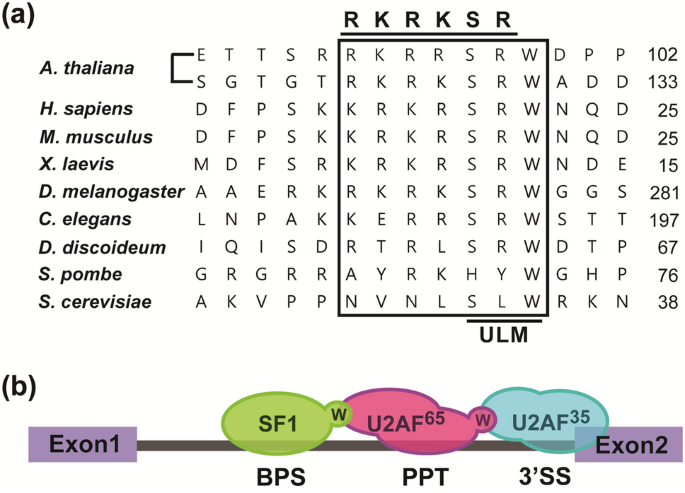
Identification of a nuclear localization signal mediating the nuclear import of Arabidopsis splicing factor1 | SpringerLink

Crystallographic Analysis of the Recognition of a Nuclear Localization Signal by the Nuclear Import Factor Karyopherin α: Cell

Crystallographic Analysis of the Recognition of a Nuclear Localization Signal by the Nuclear Import Factor Karyopherin α | Semantic Scholar

Cells | Free Full-Text | Bioinformatics and Functional Analysis of a New Nuclear Localization Sequence of the Influenza A Virus Nucleoprotein
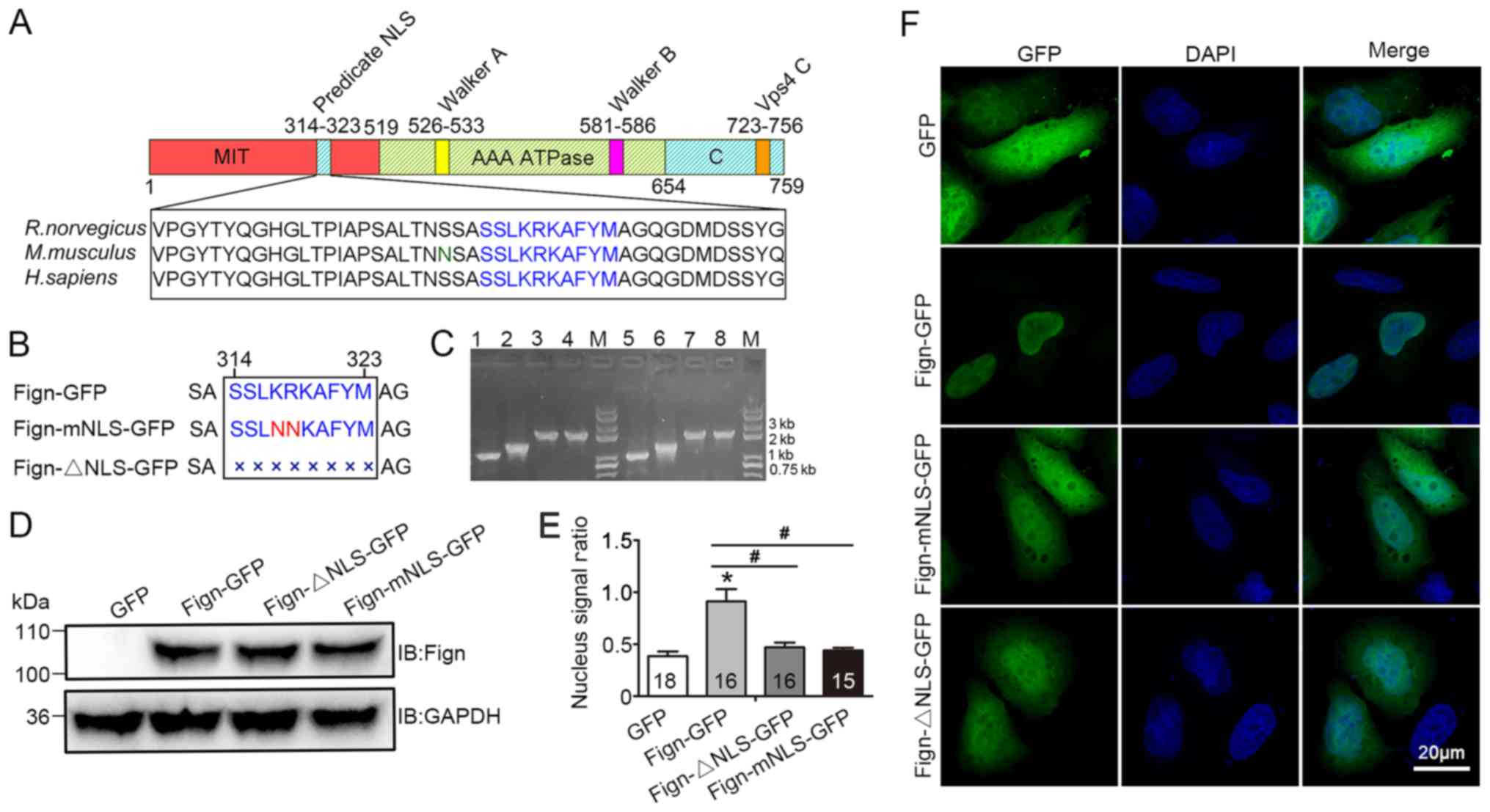
A nuclear localization signal is required for the nuclear translocation of Fign and its microtubule‑severing function

Potential NLS sequences in TPs of Streptomyces. (A) The archetypal Tpg... | Download Scientific Diagram

Table 1 from Crystallographic Analysis of the Recognition of a Nuclear Localization Signal by the Nuclear Import Factor Karyopherin α | Semantic Scholar

Cells | Free Full-Text | Bioinformatics and Functional Analysis of a New Nuclear Localization Sequence of the Influenza A Virus Nucleoprotein

The putative NLS sequence at the C-terminus of RanGAP1 is required for its nuclear accumulation in cells treated with LMB.
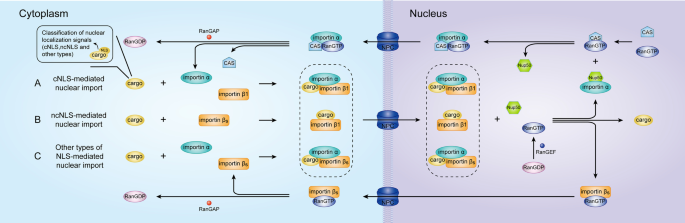
Types of nuclear localization signals and mechanisms of protein import into the nucleus | Cell Communication and Signaling | Full Text
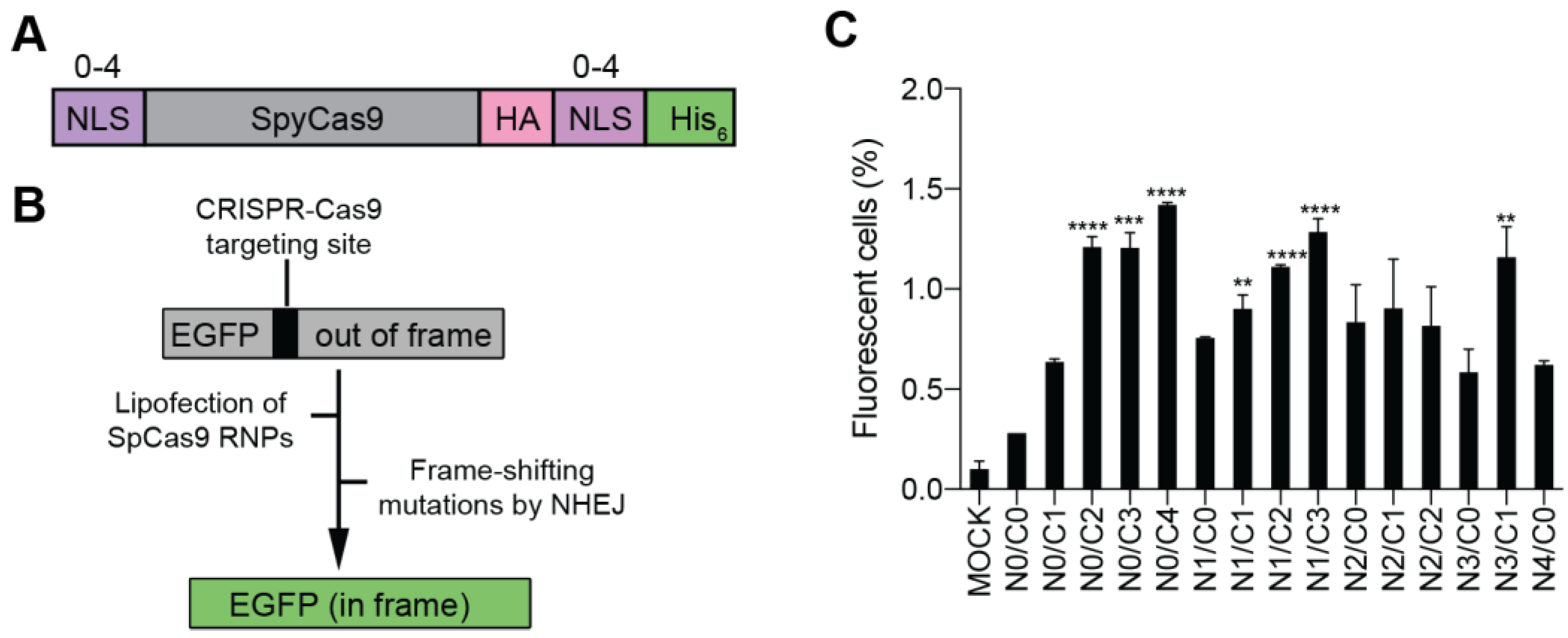
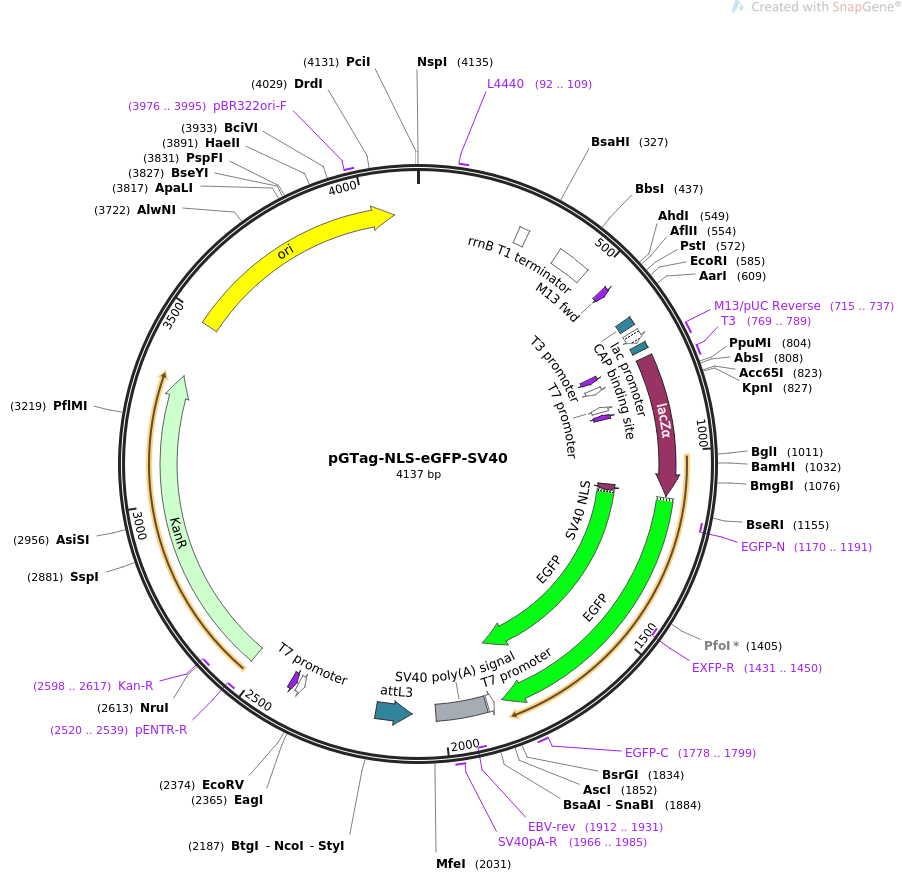
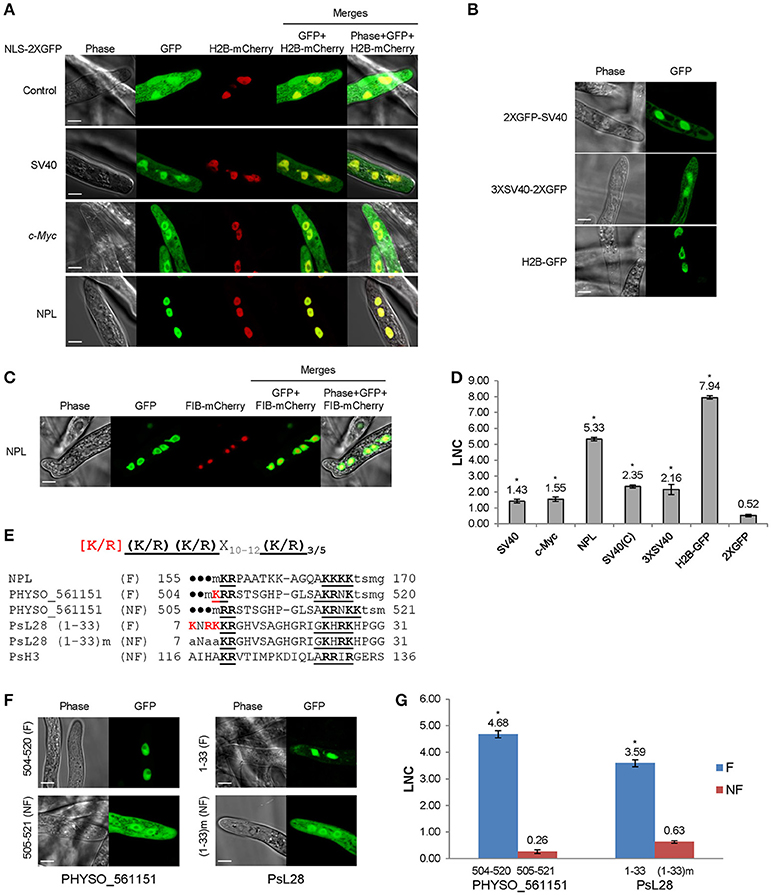
Bioengineering | Free Full-Text | Systematic Investigation of the Effects of Multiple SV40 Nuclear Localization Signal Fusion on the Genome Editing Activity of Purified SpCas9
Cooperative Nuclear Localization Sequences Lend a Novel Role to the N-Terminal Region of MSH6 | PLOS ONE

Relocalization of Nuclear Localization Sequence (NLS) and mutation in... | Download Scientific Diagram

Comparative mutagenesis of nuclear localization signals reveals the importance of neutral and acidic amino acids | Semantic Scholar