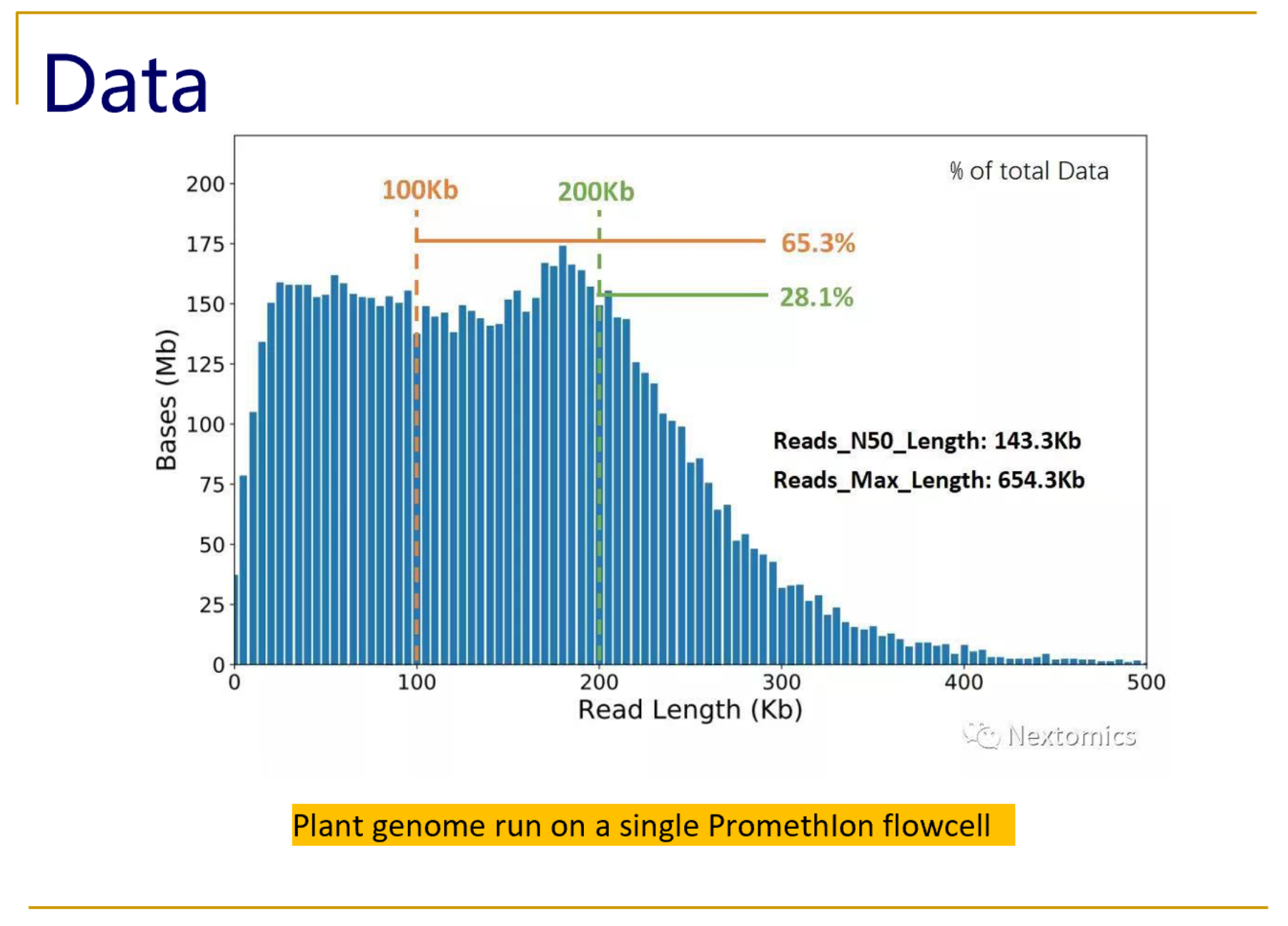
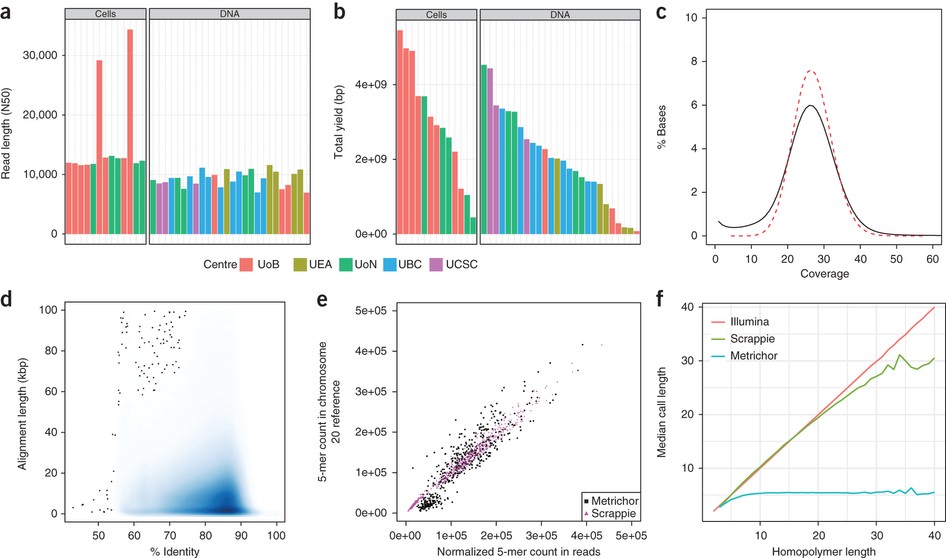
Oxford Nanopore Technology: A Promising Long-Read Sequencing Platform To Study Exon Connectivity and Characterize Isoforms of Complex Genes | Semantic Scholar

Nanopore sequencing: Review of potential applications in functional genomics - Kono - 2019 - Development, Growth & Differentiation - Wiley Online Library
![Evaluation of full-length nanopore 16S sequencing for detection of pathogens in microbial keratitis [PeerJ] Evaluation of full-length nanopore 16S sequencing for detection of pathogens in microbial keratitis [PeerJ]](https://dfzljdn9uc3pi.cloudfront.net/2021/10778/1/fig-1-full.png)
Evaluation of full-length nanopore 16S sequencing for detection of pathogens in microbial keratitis [PeerJ]

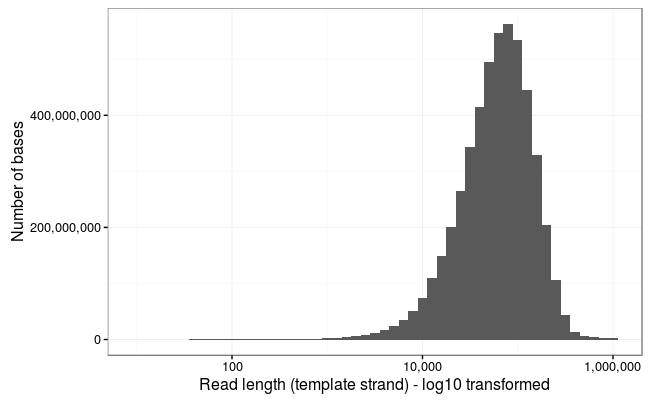
From kilobases to "whales": a short history of ultra-long reads and high-throughput genome sequencing
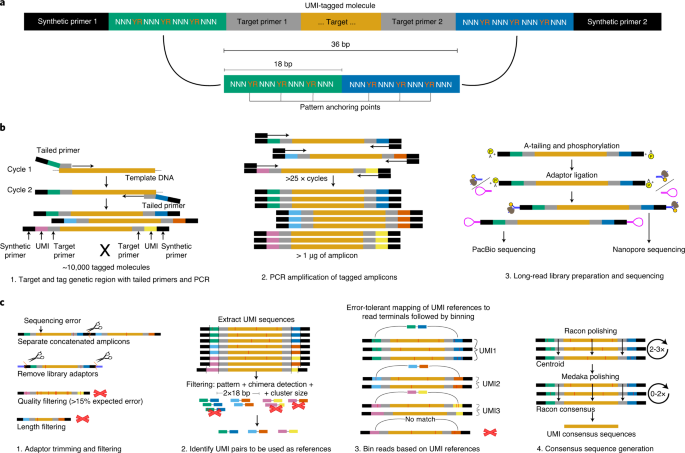
High-accuracy long-read amplicon sequences using unique molecular identifiers with Nanopore or PacBio sequencing | Nature Methods