Evaluation of ultra-low input RNA sequencing for the study of human T cell transcriptome | Scientific Reports

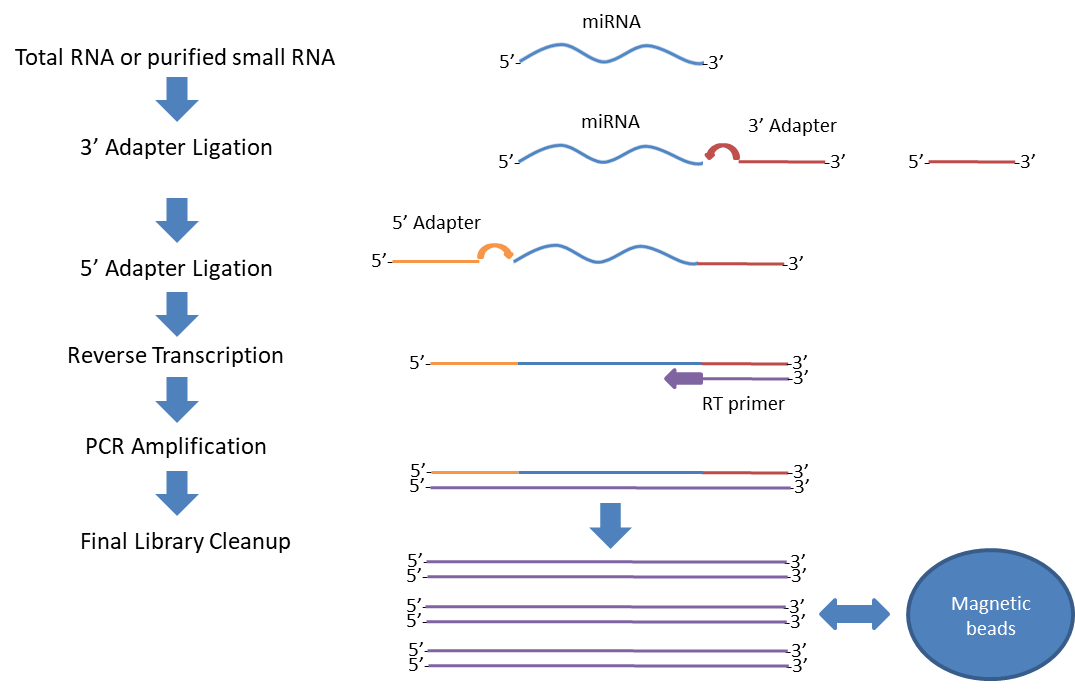
High-throughput, Efficient, and Unbiased Capture of Small RNAs from Low- input Samples for Sequencing | Scientific Reports
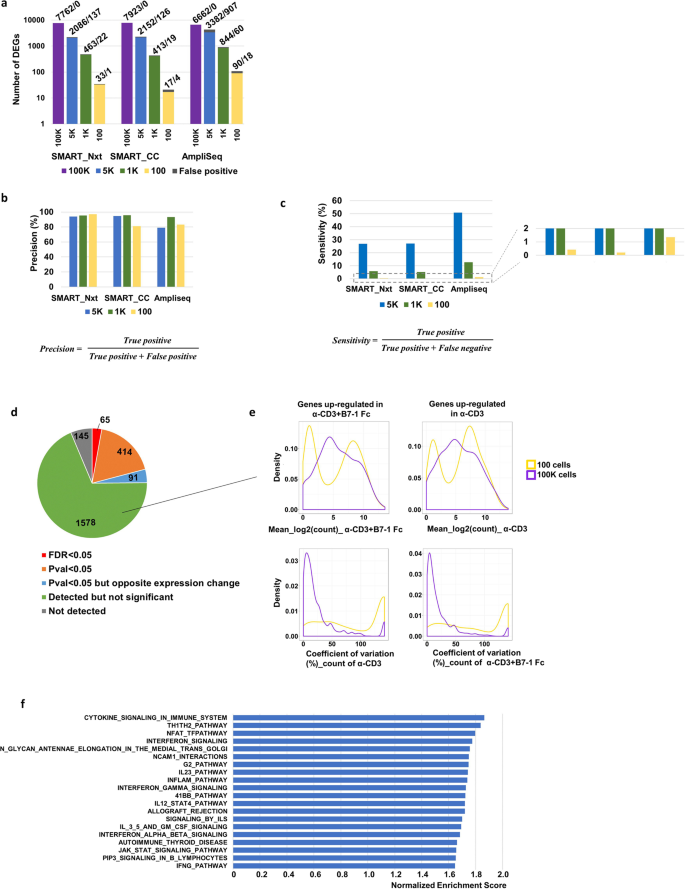
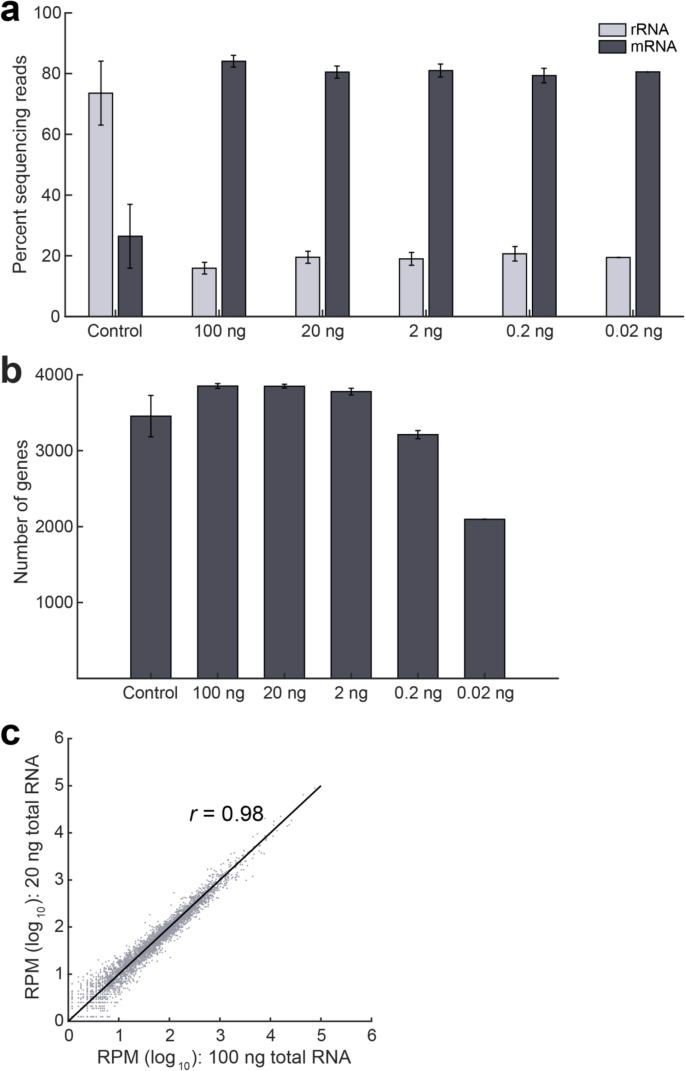
Evaluation of ultra-low input RNA sequencing for the study of human T cell transcriptome | Scientific Reports
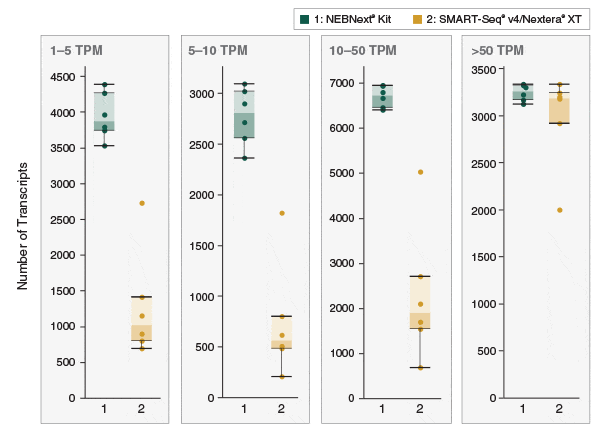
Erstklassige RNA-Sequenzierlibraries aus Einzelzellen – NEBNext Single Cell/Low Input RNA Library Prep - New England Biolabs GmbH

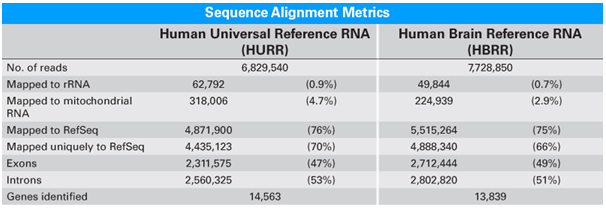
Protocol for RNA-seq library preparation starting from a rare muscle stem cell population or a limited number of mouse embryonic stem cells - ScienceDirect
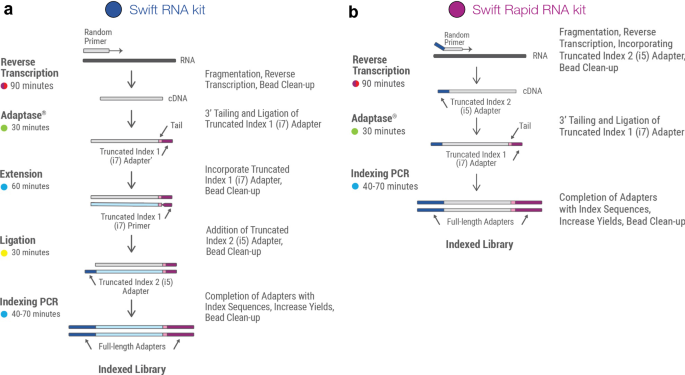
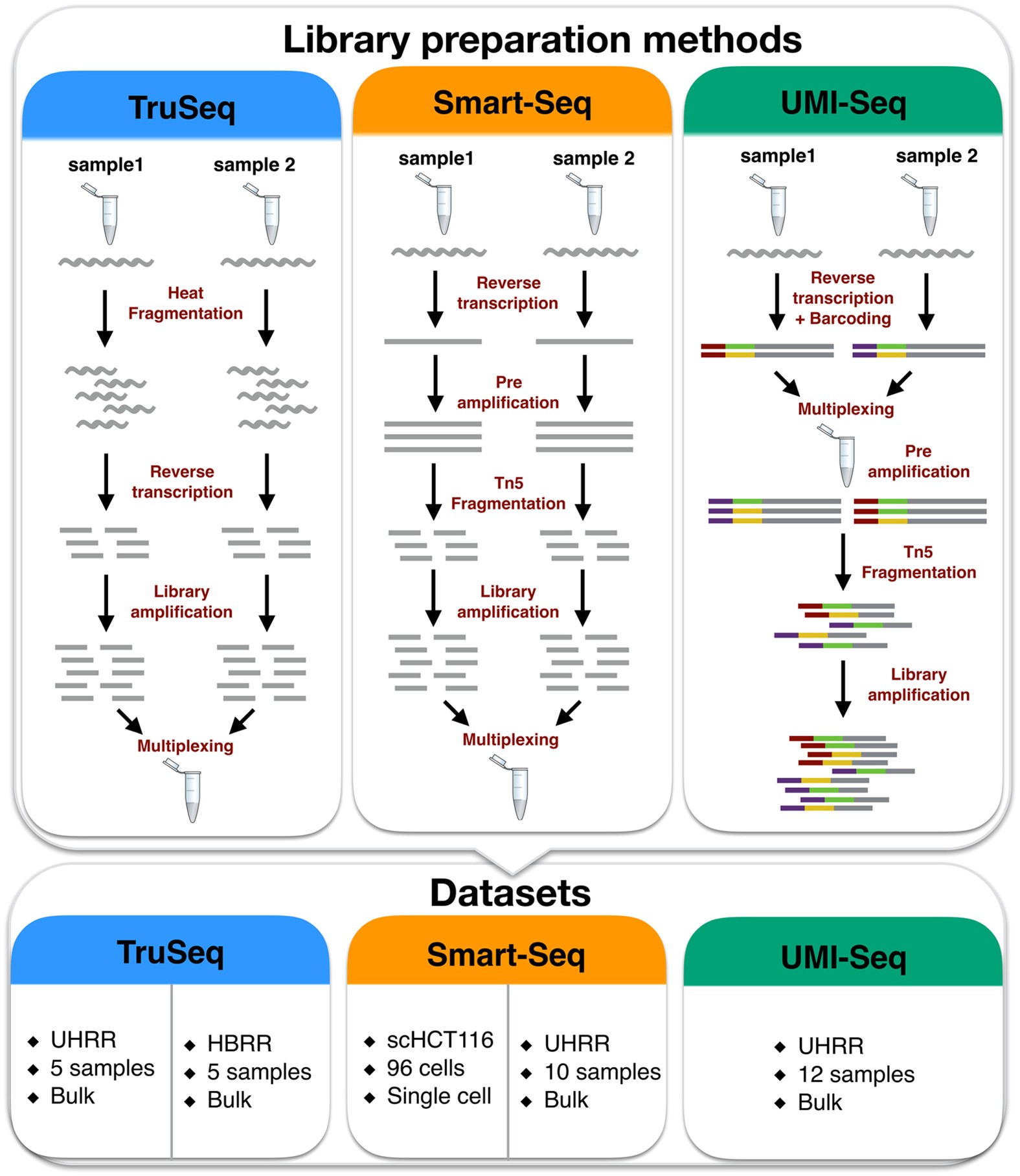
Systematic comparative analysis of strand-specific RNA-seq library preparation methods for low input samples | Scientific Reports
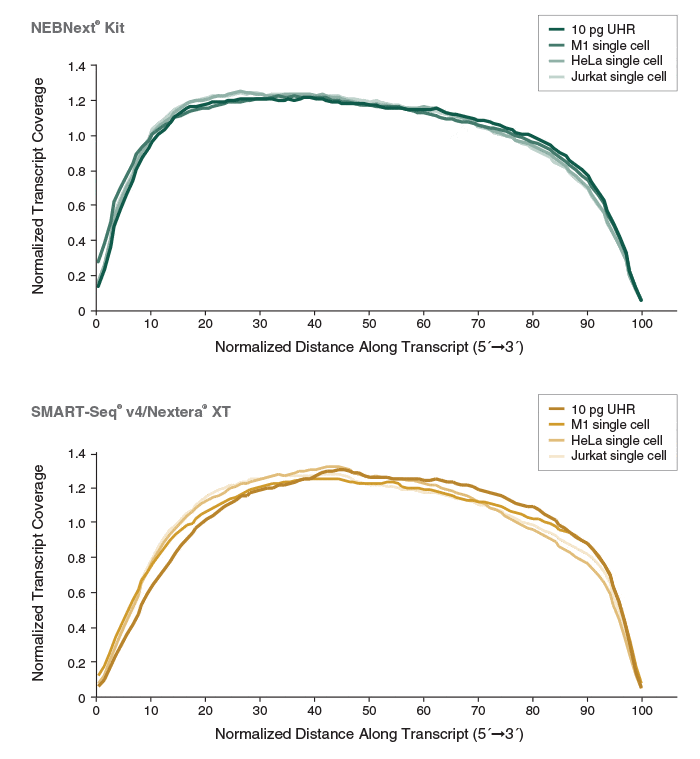
Erstklassige RNA-Sequenzierlibraries aus Einzelzellen – NEBNext Single Cell/Low Input RNA Library Prep - New England Biolabs GmbH
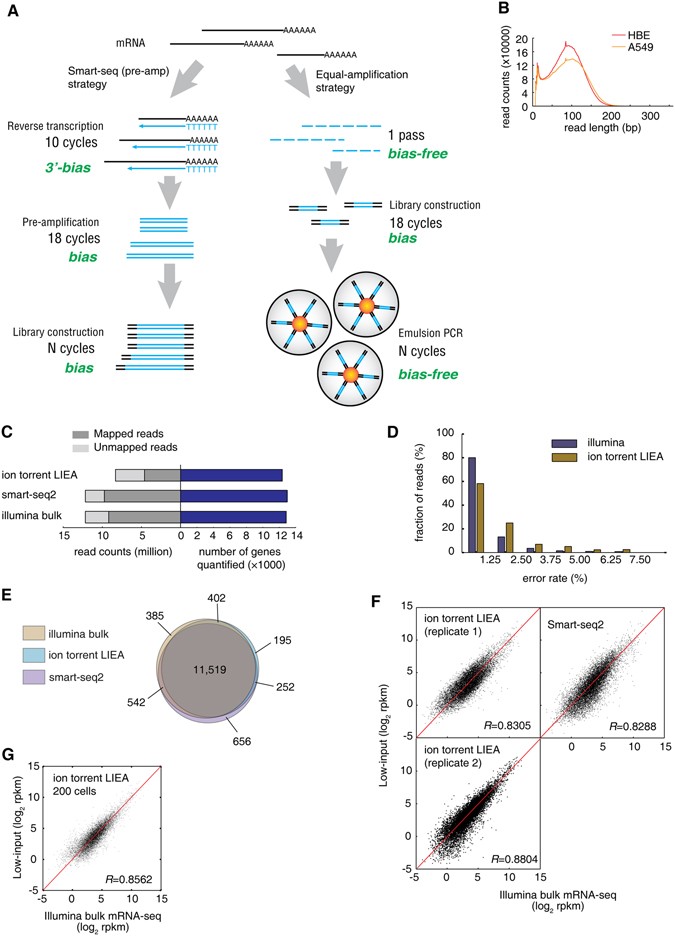
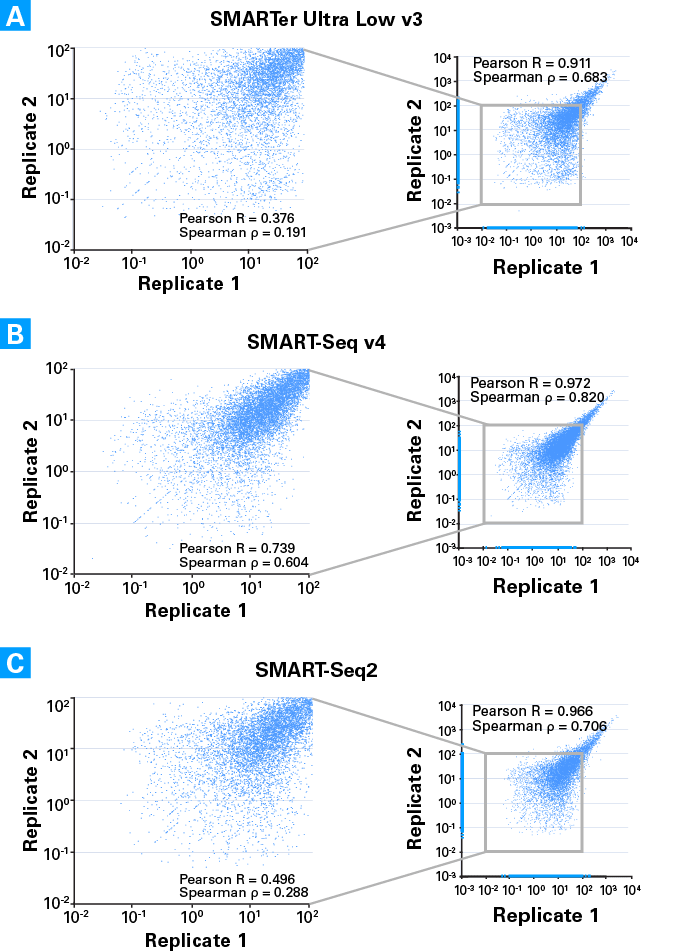
Low-cost, Low-bias and Low-input RNA-seq with High Experimental Verifiability based on Semiconductor Sequencing | Scientific Reports