Interplay between insert size, GC content and sequencing quality of a... | Download Scientific Diagram

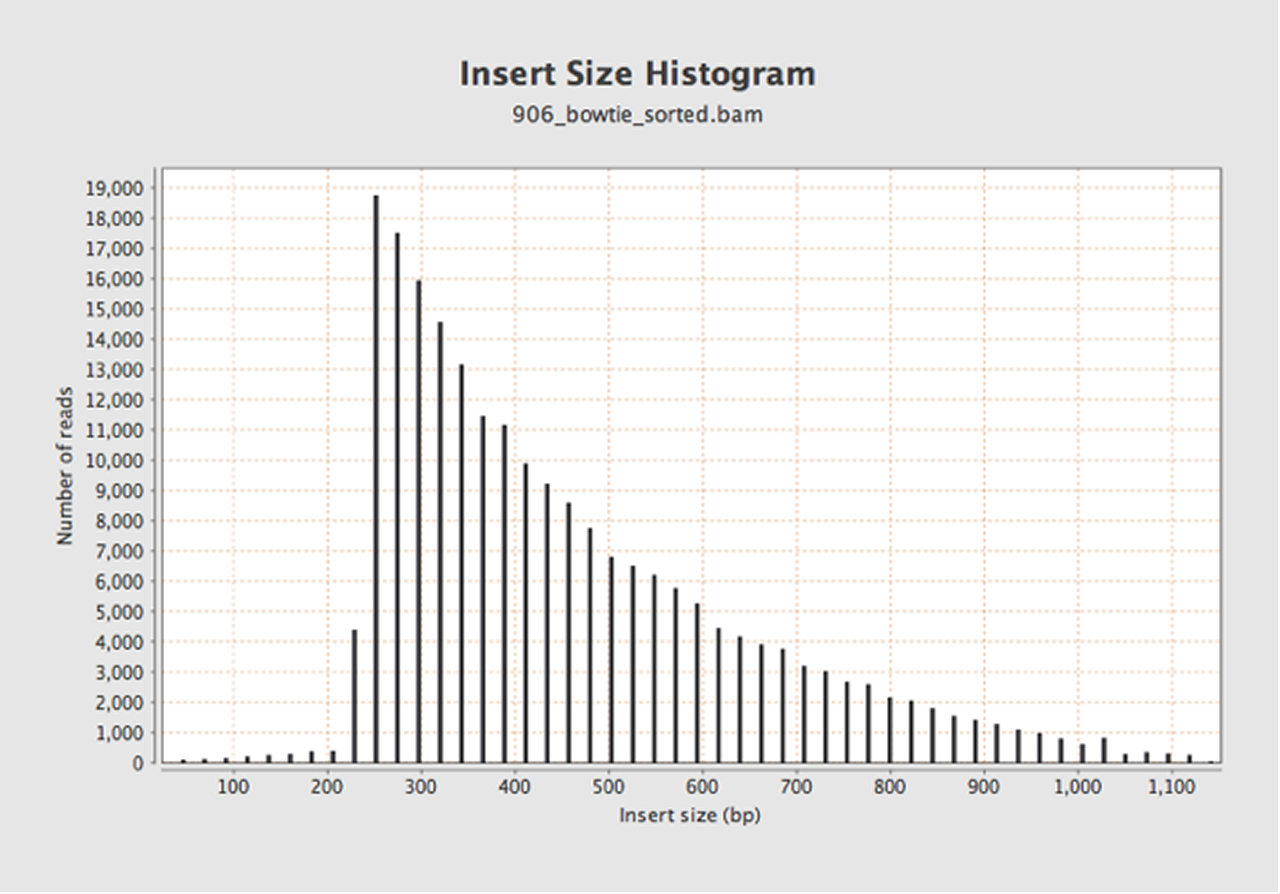
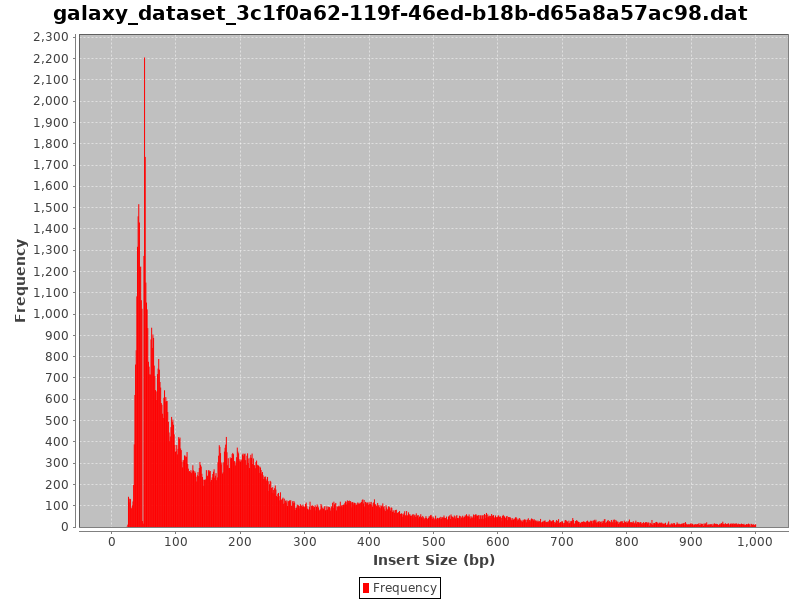
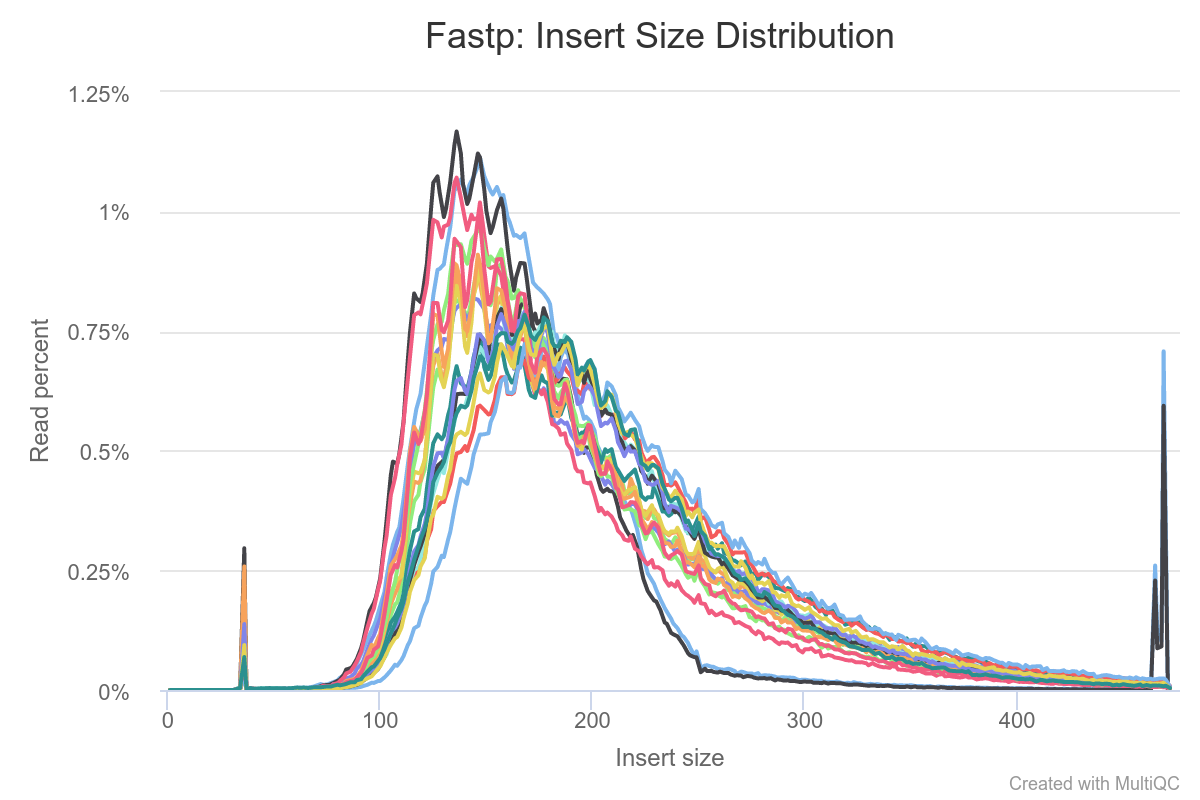
Insert size and (b) GC profiles of Illumina sequence data from each of... | Download Scientific Diagram

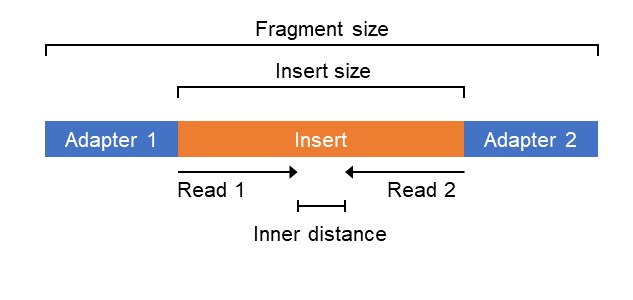
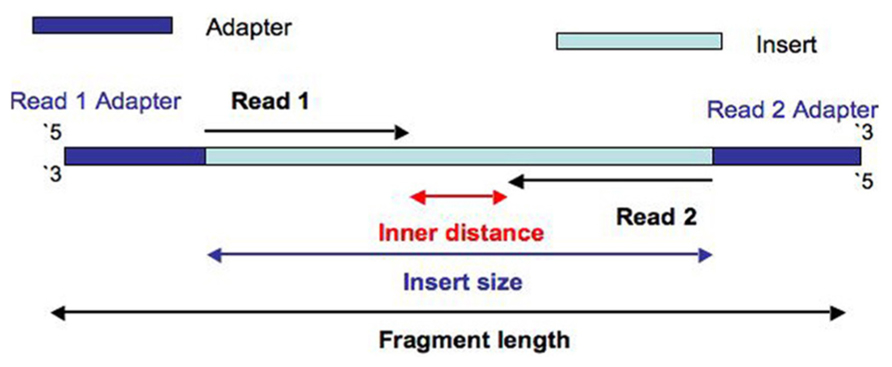
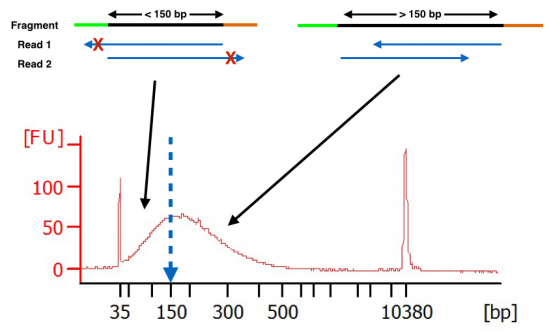
Definition of essential terms. (a) Definition of fragment, read, and... | Download Scientific Diagram
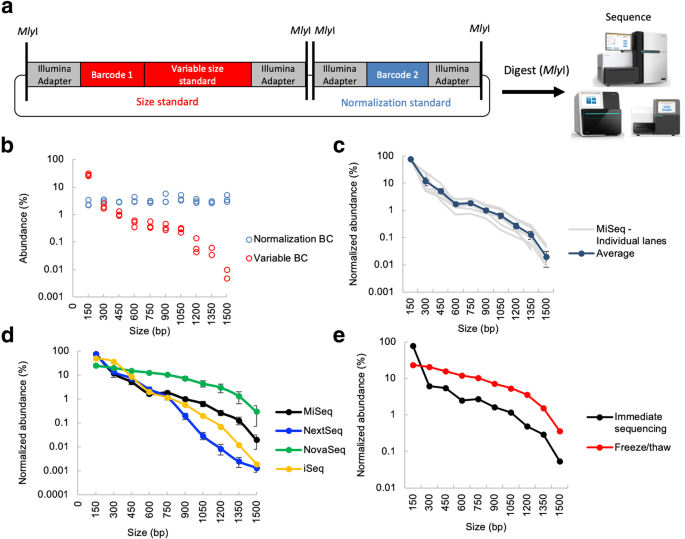
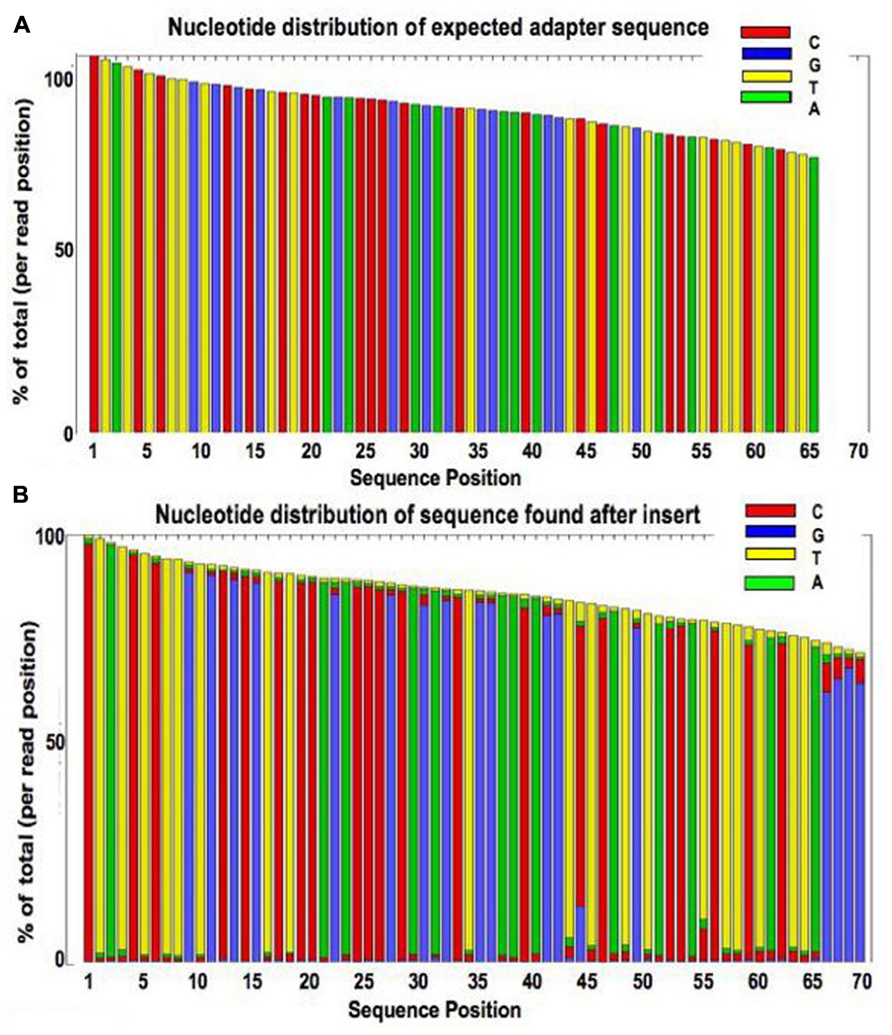
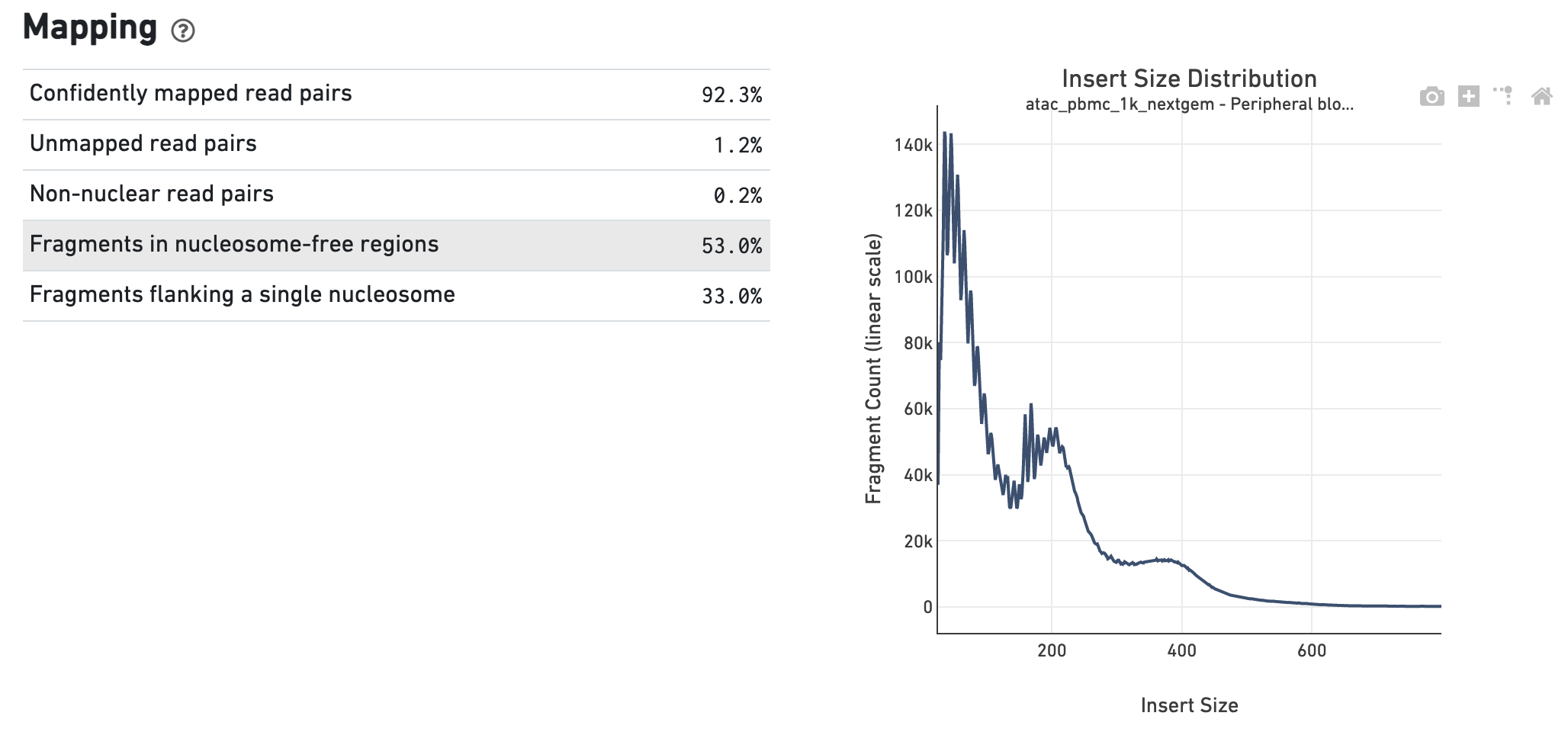
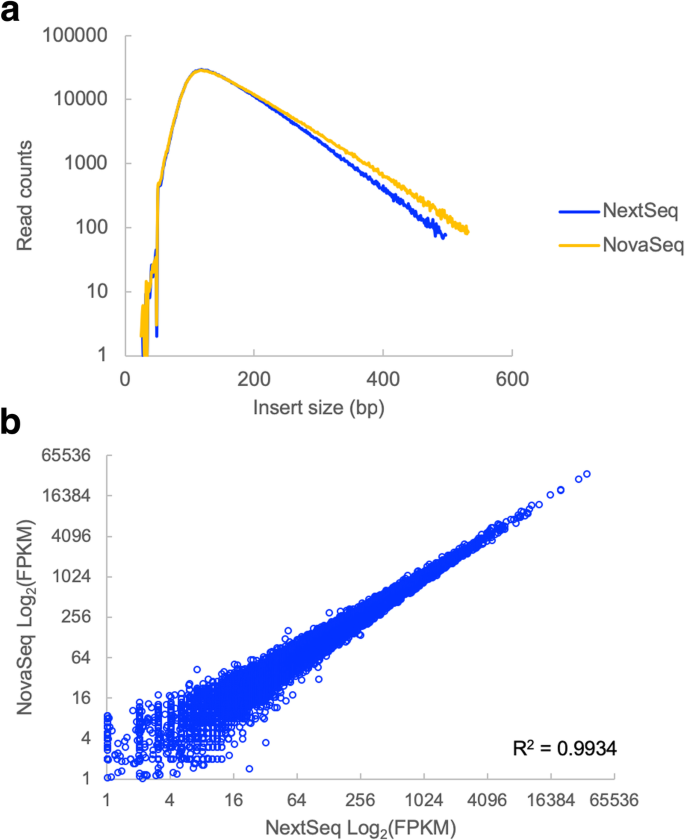
Measuring sequencer size bias using REcount: a novel method for highly accurate Illumina sequencing-based quantification | Genome Biology | Full Text
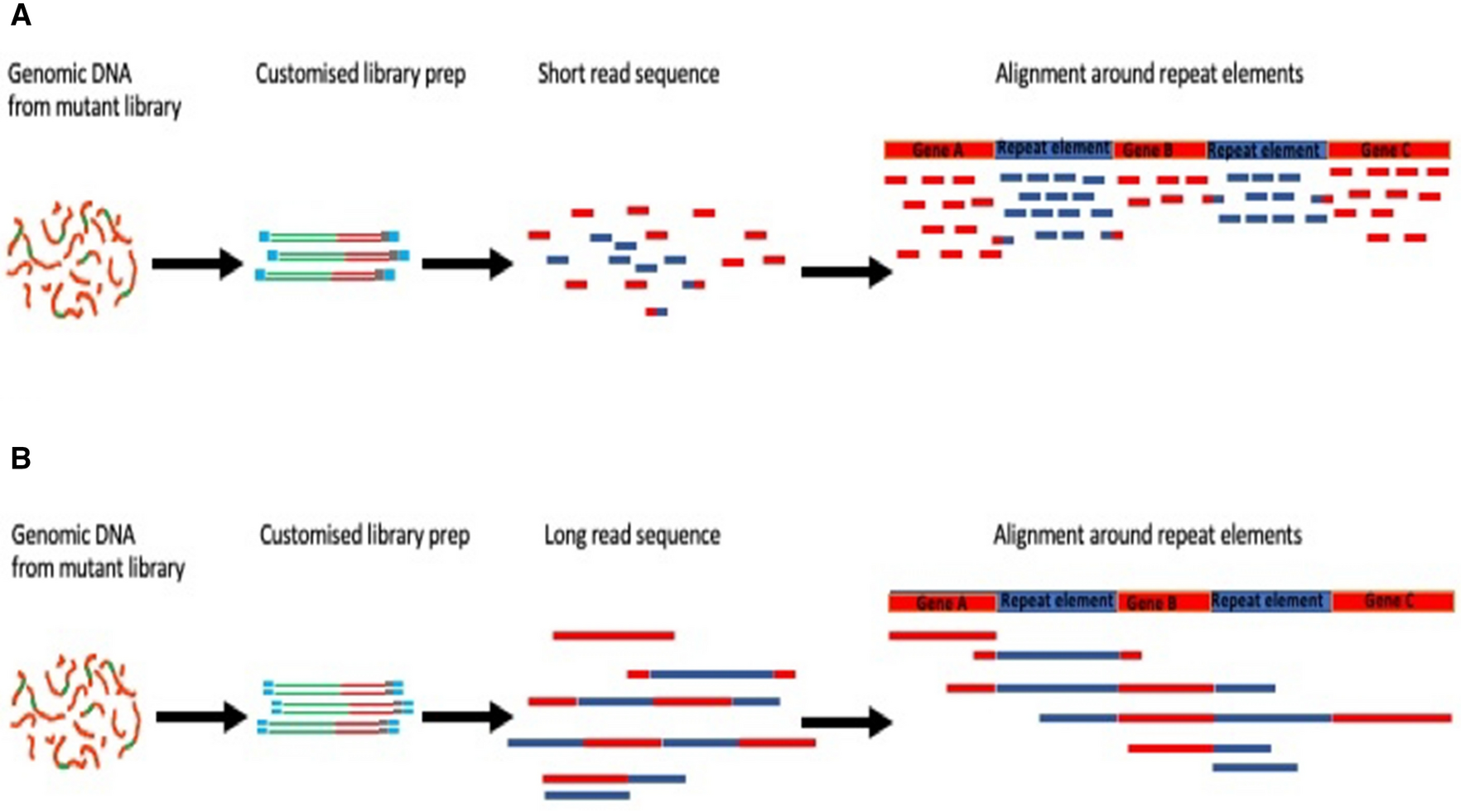
Long-read sequencing for identification of insertion sites in large transposon mutant libraries | Scientific Reports

CUT&Tag-BS: an efficient and low-cost method for simultaneous profiling of histone modification and DNA methylation | bioRxiv

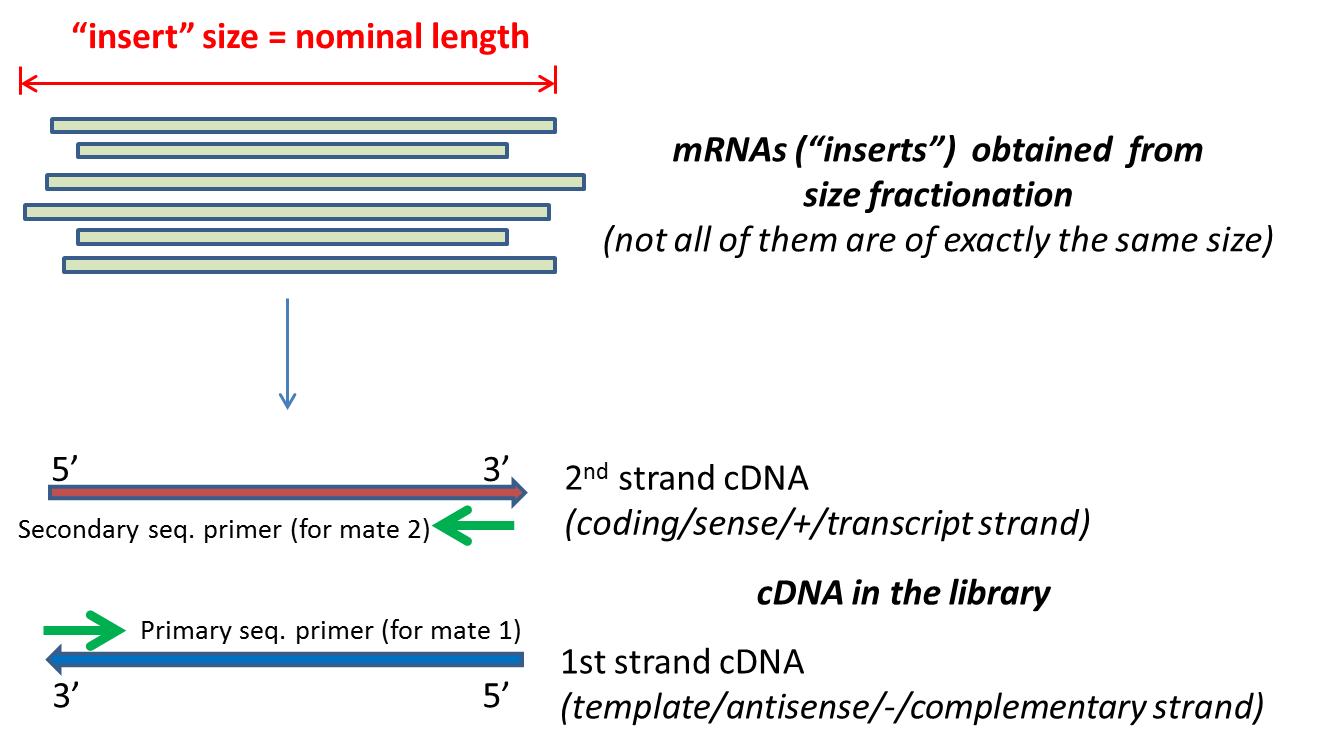
Compbio 020: Reads, fragments and inserts - what you need to know for understanding your sequencing data — Bad Grammar, Good Syntax

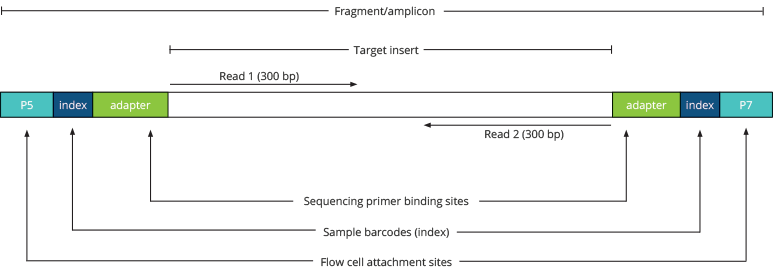
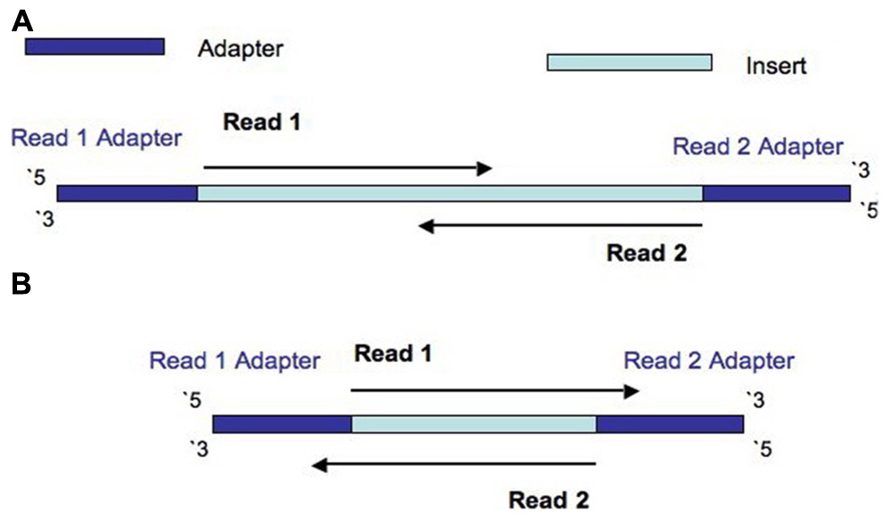
Library insert size. Libraries were constructed using the standard (A)... | Download Scientific Diagram
Measuring sequencer size bias using REcount: a novel method for highly accurate Illumina sequencing-based quantification | Genome Biology | Full Text