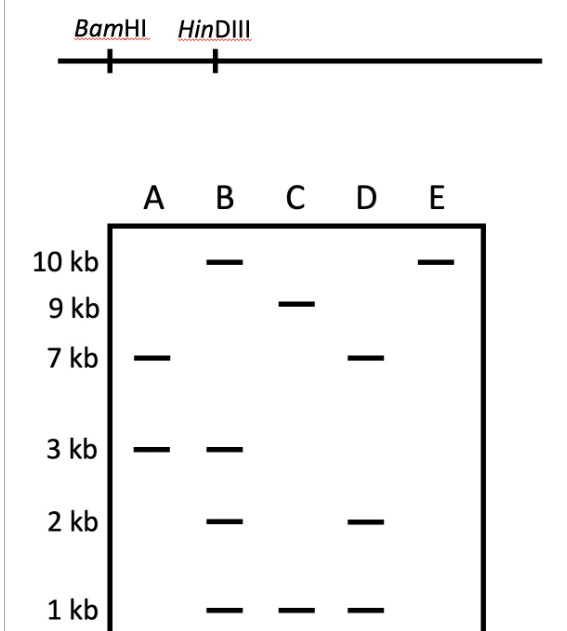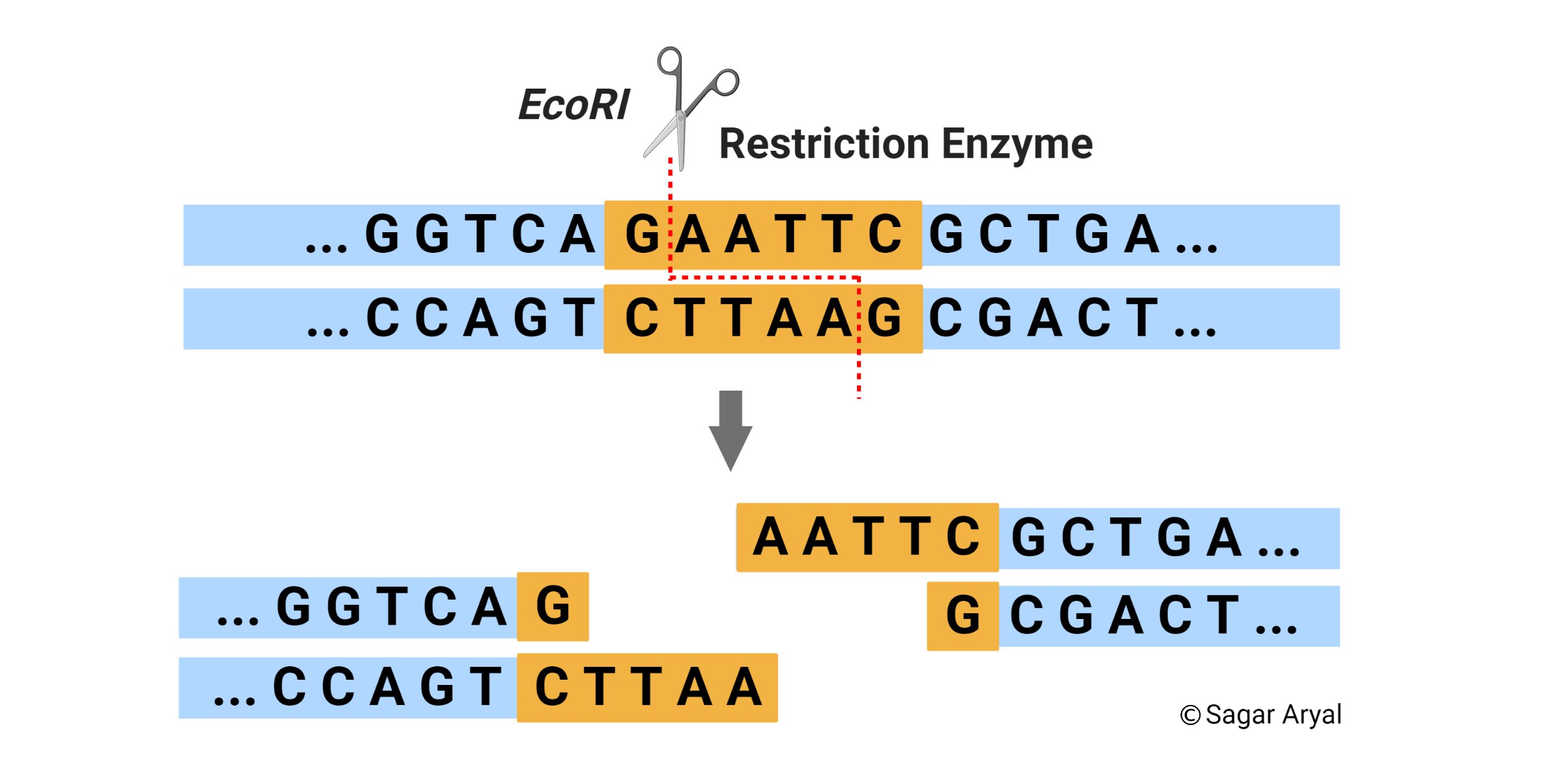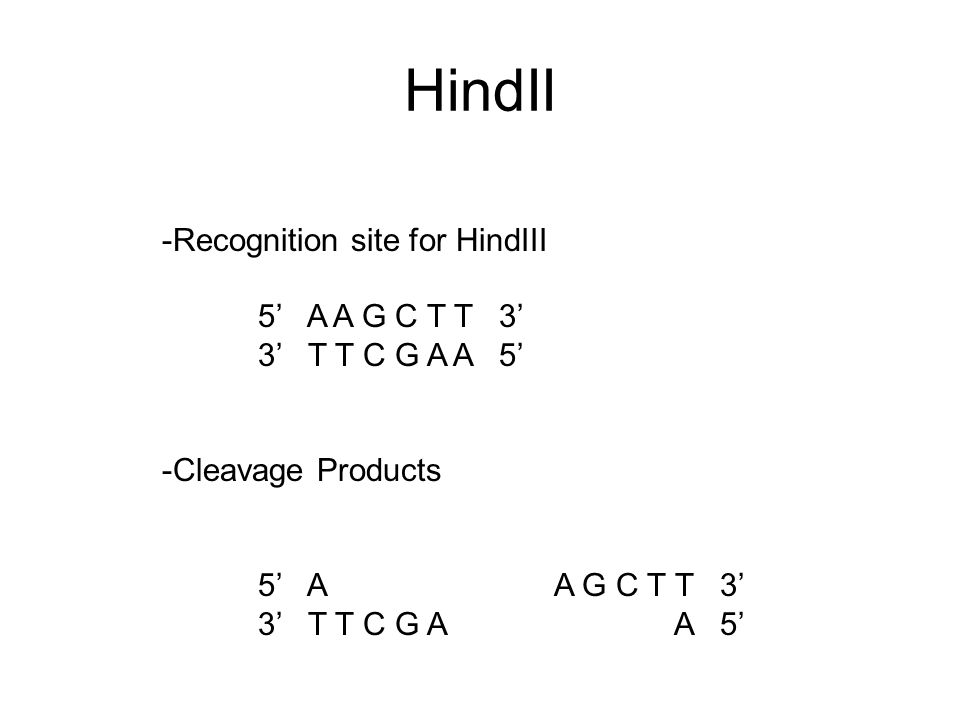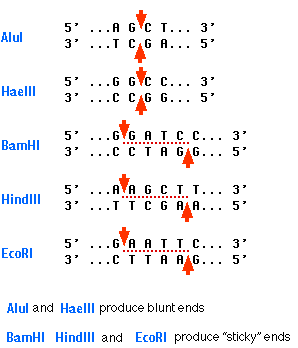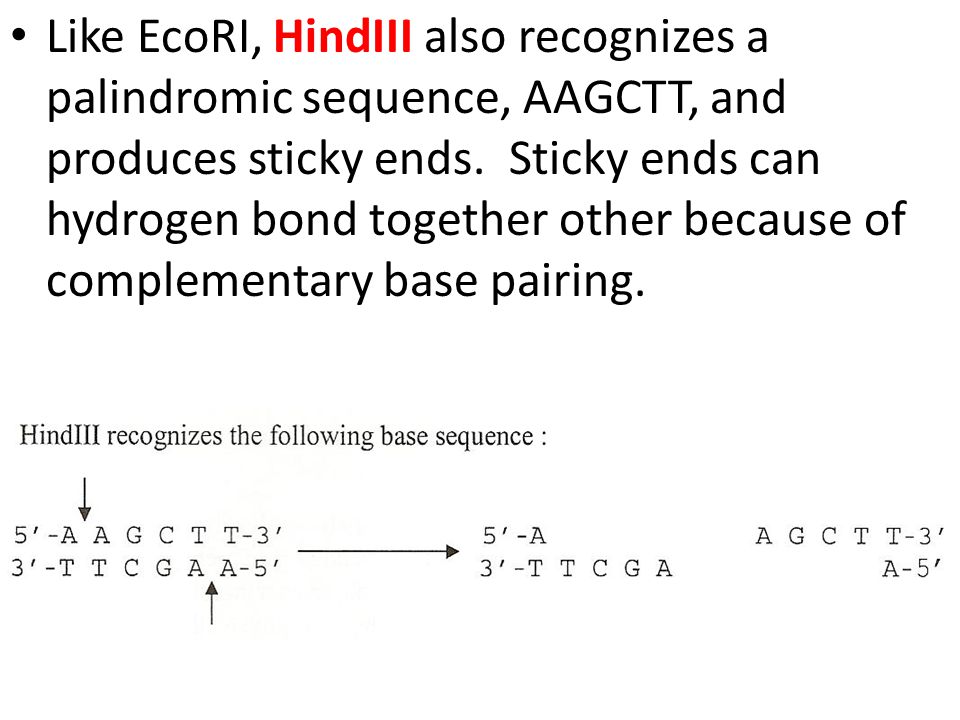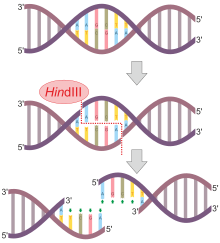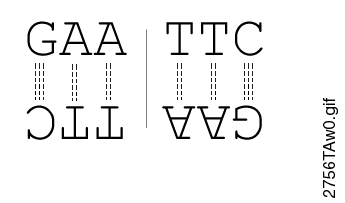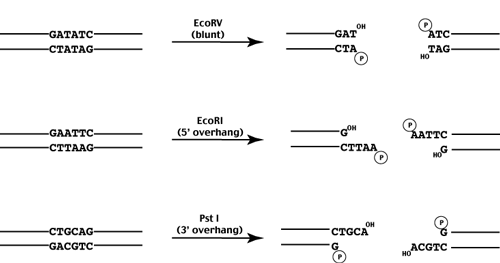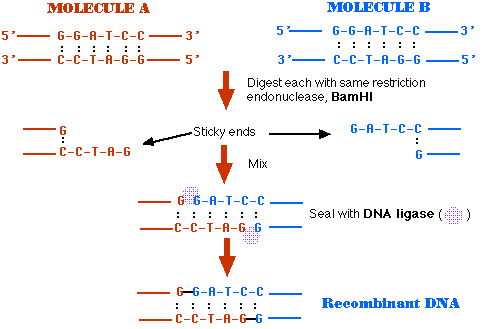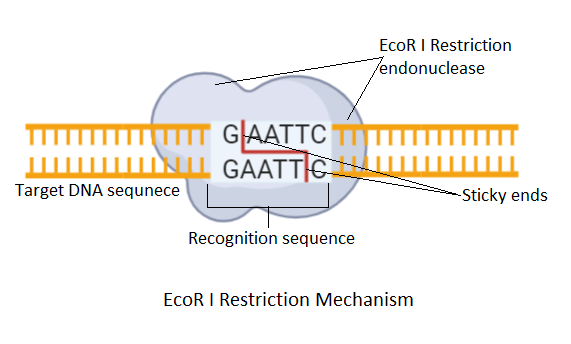
GAATTC is the restriction site, for which of the following restriction endonucleases?A. HindIIIB. EcoRIC. BamID. HaeIII

Plasmids used in the study. A 1.5-kb HindIII URA3 fragment was cloned... | Download Scientific Diagram

Problem‐solving test: Restriction endonuclease mapping - Szeberényi - 2011 - Biochemistry and Molecular Biology Education - Wiley Online Library
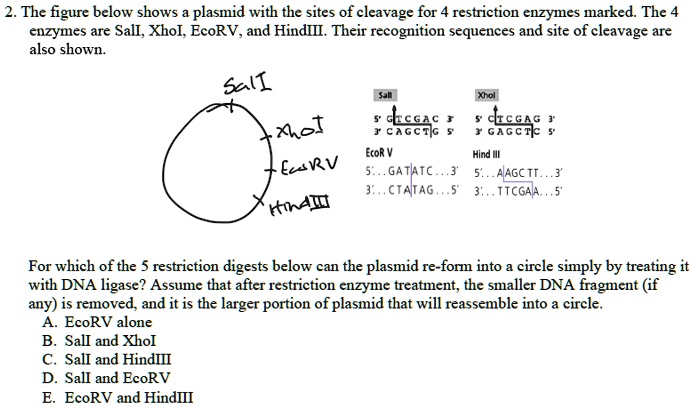
SOLVED: 2. The figure ' below shows plasmid with the sites of cleavage for restriction enzymes marked. The enzymes are Sall, Xhol, EcoRV, and HindIII Their recognition sequences and site of cleavage
Useful tool to generate unidirectional deletion vectors by utilizing the star activity of BamHI in an NcoI-BamHI-XhoI cassette | BioTechniques