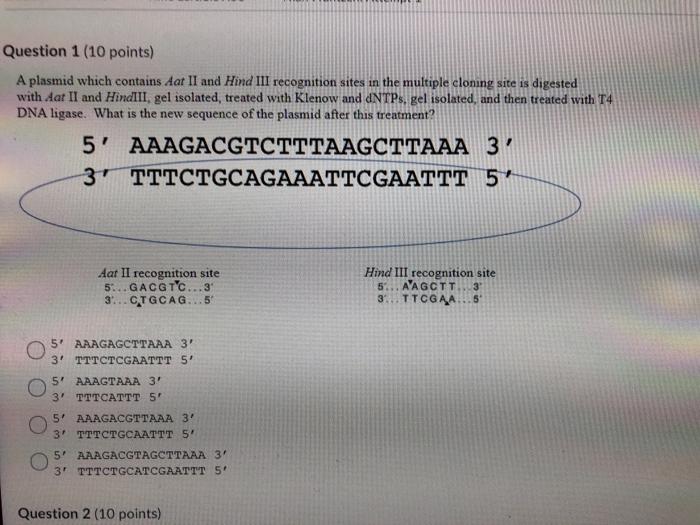
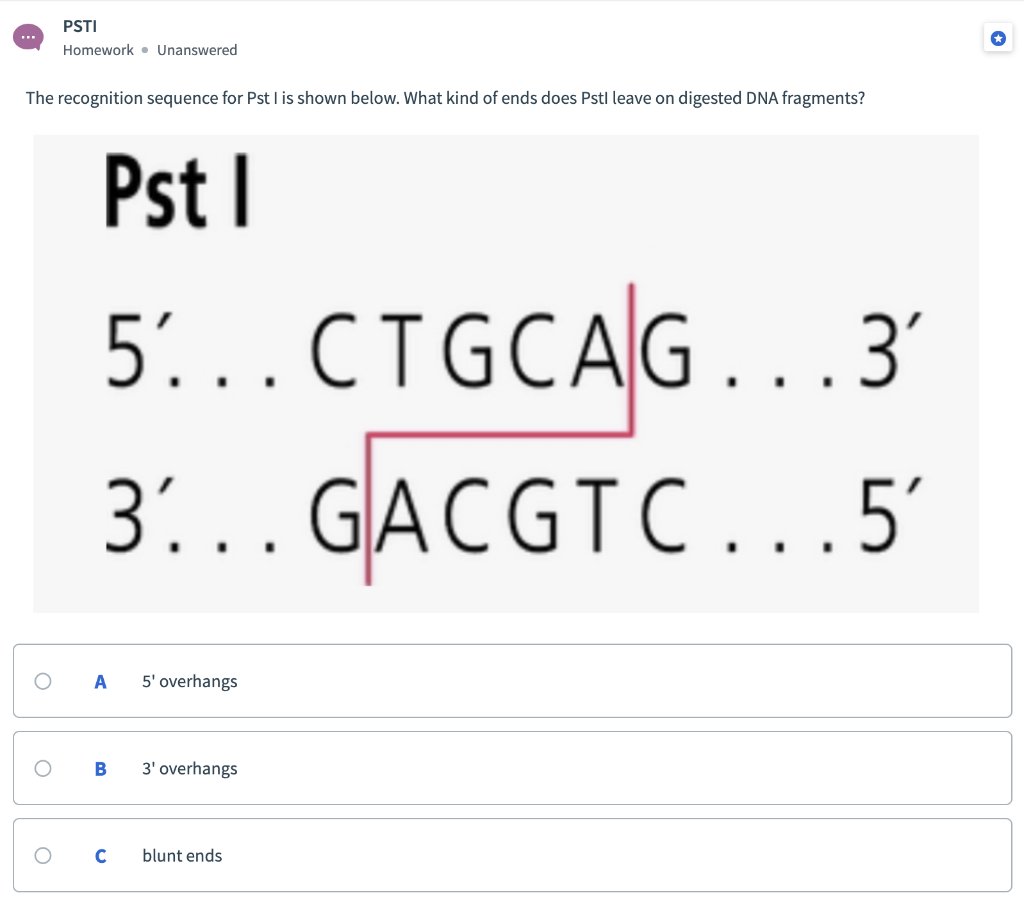
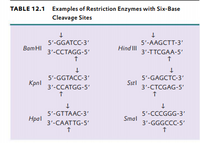
Restriction Enzymes/endonucleases Restriction enzymes are proteins that scan the DNA for specific sequences, usually palindromic sequences of 4 to 8 nucleotides, - ppt download

Recognition sequences Restriction enzyme EcoRI cuts the DNA into fragments. Sticky end. - ppt download

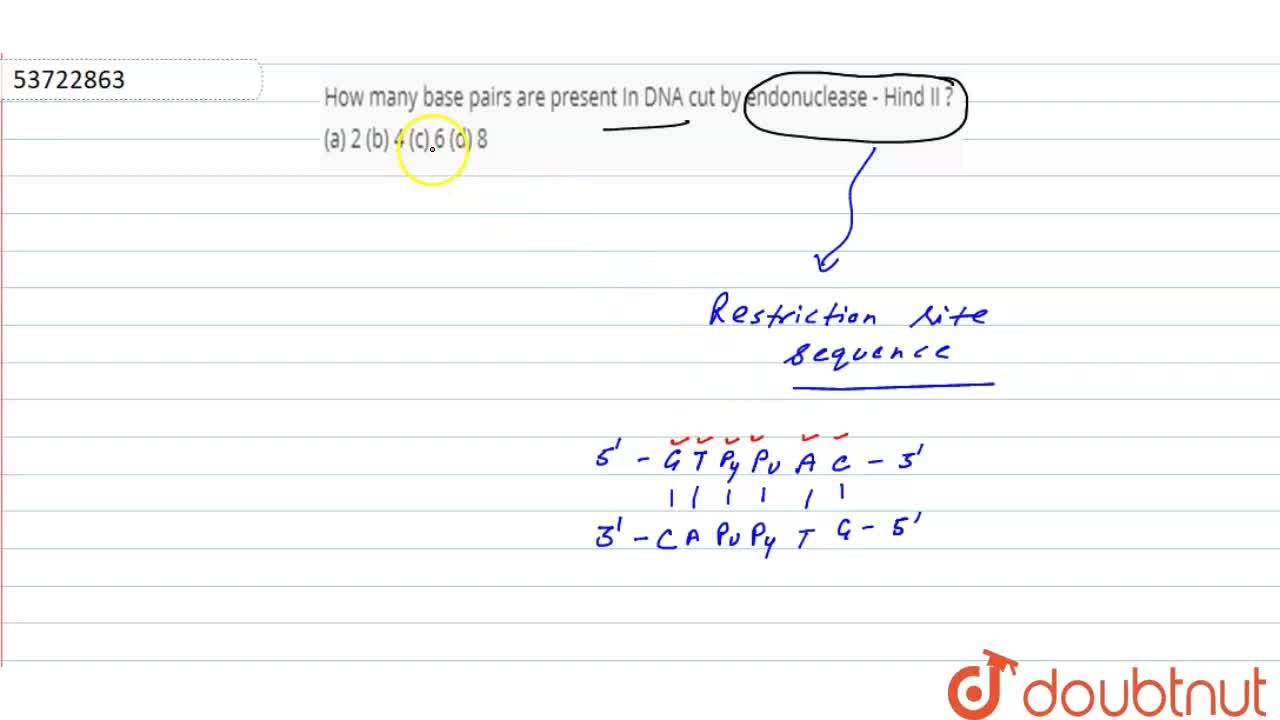
Restriction endonuclease - Hind II always cuts DNA molecules at a particular point by recognizin... - YouTube

Trick to Learn Restriction Site| Biotech Tricks NEET | Biotech mnemonic | Biotechnology Tricks NEET - YouTube
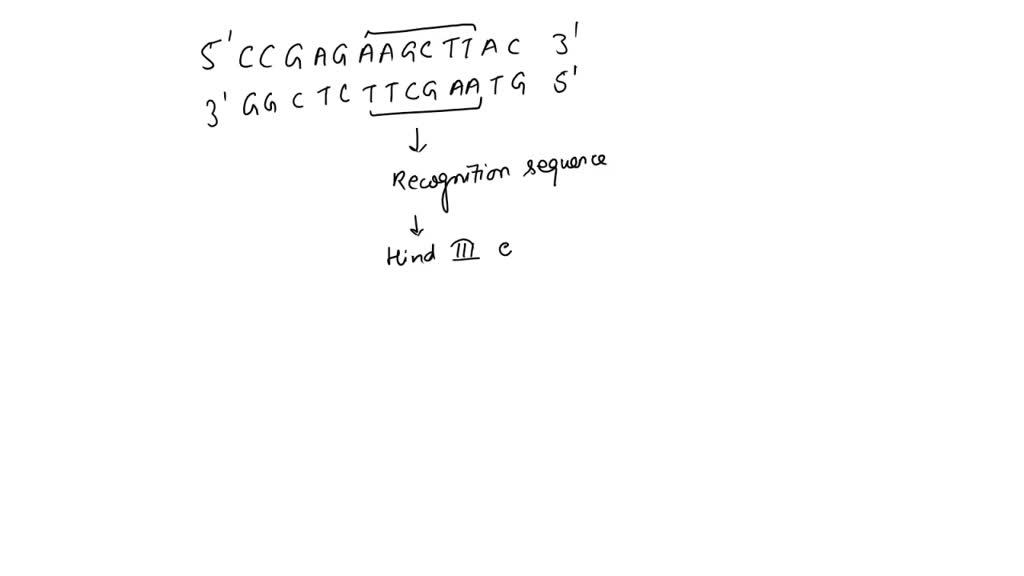
SOLVED: The DNA sequence (5 '-CCGAGAGATCTAC-3') contains a six-base sequence that is a recognition and cutting site for a restriction enzyme. ( 4 points) (a) What is this sequence? (2 points) (b)

It was found that Hind 2 always cuts DNA molecules at a particular point by recognising a specific sequence - Brainly.in

What is the Difference Between EcoRI and HindIII Restriction Enzymes | Compare the Difference Between Similar Terms
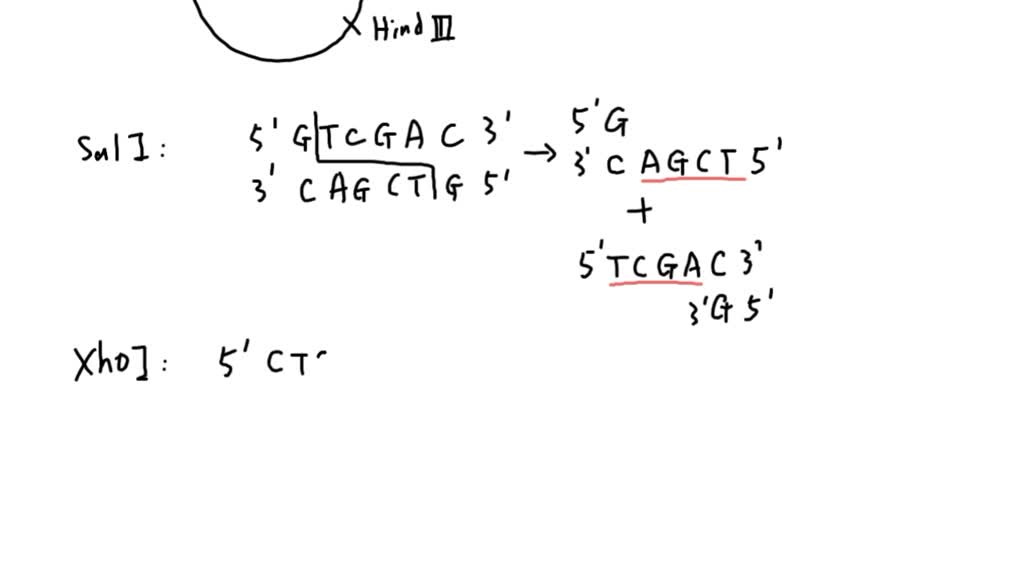
SOLVED: 2. The figure ' below shows plasmid with the sites of cleavage for restriction enzymes marked. The enzymes are Sall, Xhol, EcoRV, and HindIII Their recognition sequences and site of cleavage

SOLVED: 'If the recognition sequence of the restriction enzyme Hindlll is AAGCTT, then how many covalent bonds will be broken by the enzyme in the following DNA molecule? STCAAGCTCC-G-A-A-G-CTTGA JAGTSTCGAAGCTTC-GAACT 5 8.2

Which among the following restriction endonucleases will produce blunt ends?I. BamH III. Hind IIIII. EcoR IIV. Hind IIIV. Sal I

The restriction enzyme responsible for the cleavage of following sequence is 5' - G - T - C - G - A - C - 3'3' - C - A - G - C - T - G - 5'
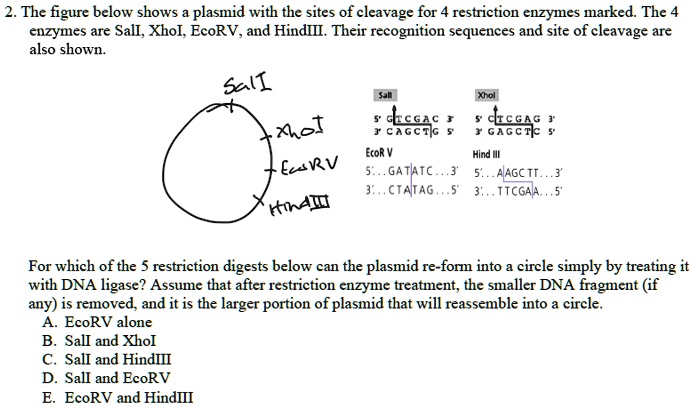
SOLVED: 2. The figure ' below shows plasmid with the sites of cleavage for restriction enzymes marked. The enzymes are Sall, Xhol, EcoRV, and HindIII Their recognition sequences and site of cleavage