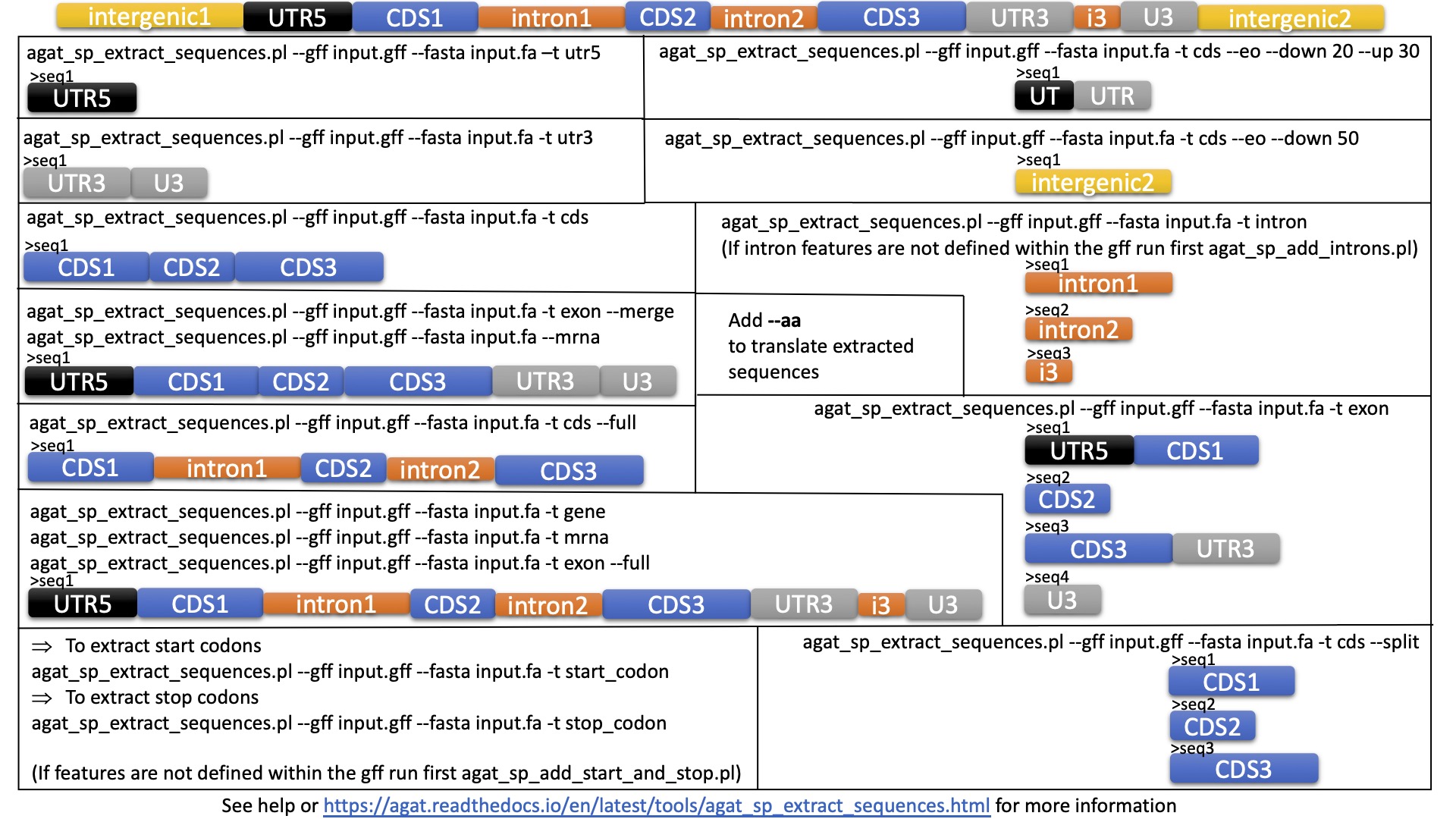
GitHub - IdoBar/FastRetriever: A python script used to extract specific sequences from a fasta file based on ID or keyword
GitHub - anatolydryga/FilterFasta: filter FASTA file based on sequence identifier and extract subsequences from start to end position
Get the parent gene ID when extracting fasta sequences with a gff file. · Issue #210 · shenwei356/seqkit · GitHub