Overview of the IRFinder algorithm. QC quality control. ERCC Sequences... | Download Scientific Diagram
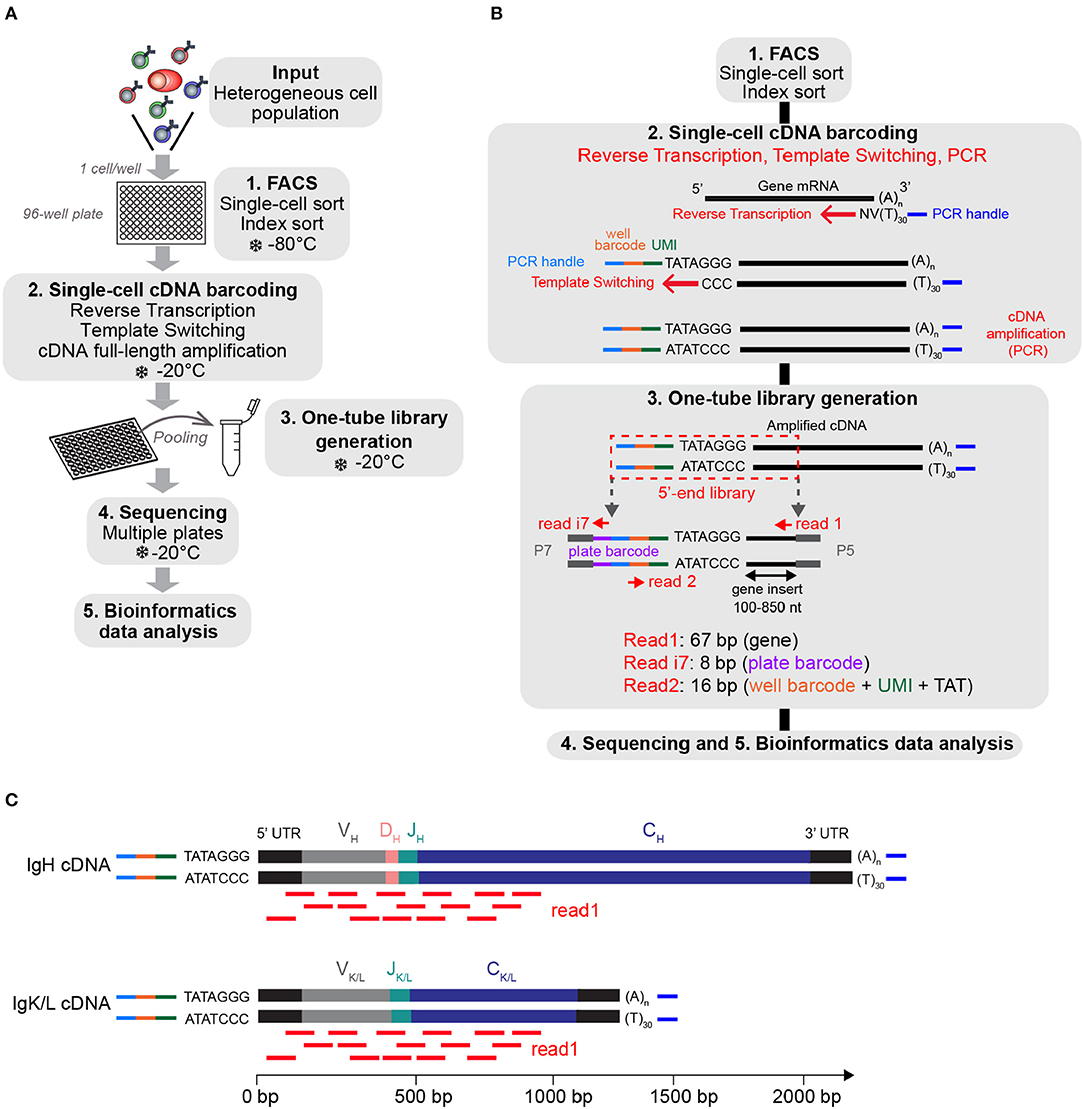
Frontiers | FB5P-seq: FACS-Based 5-Prime End Single-Cell RNA-seq for Integrative Analysis of Transcriptome and Antigen Receptor Repertoire in B and T Cells
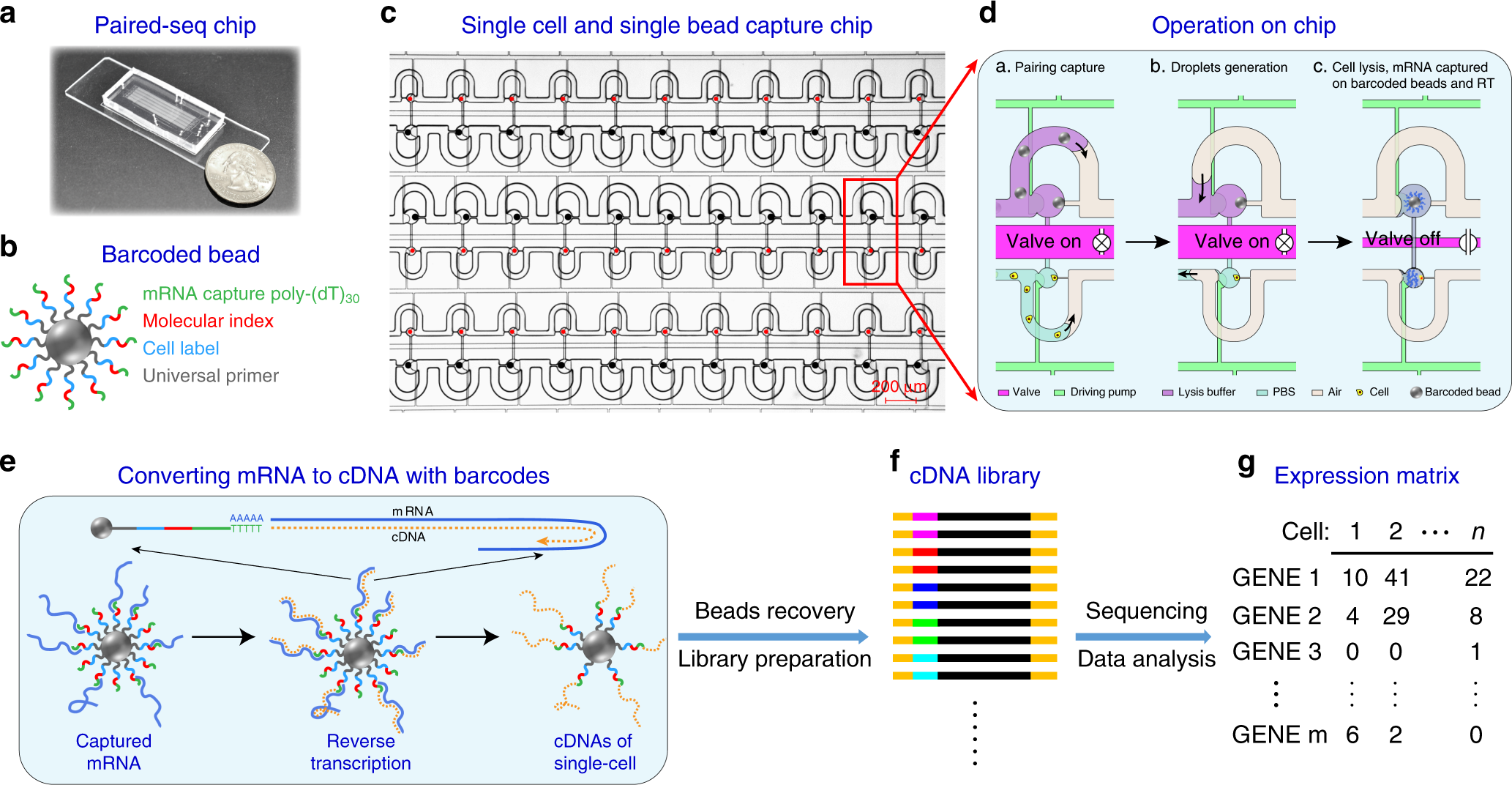
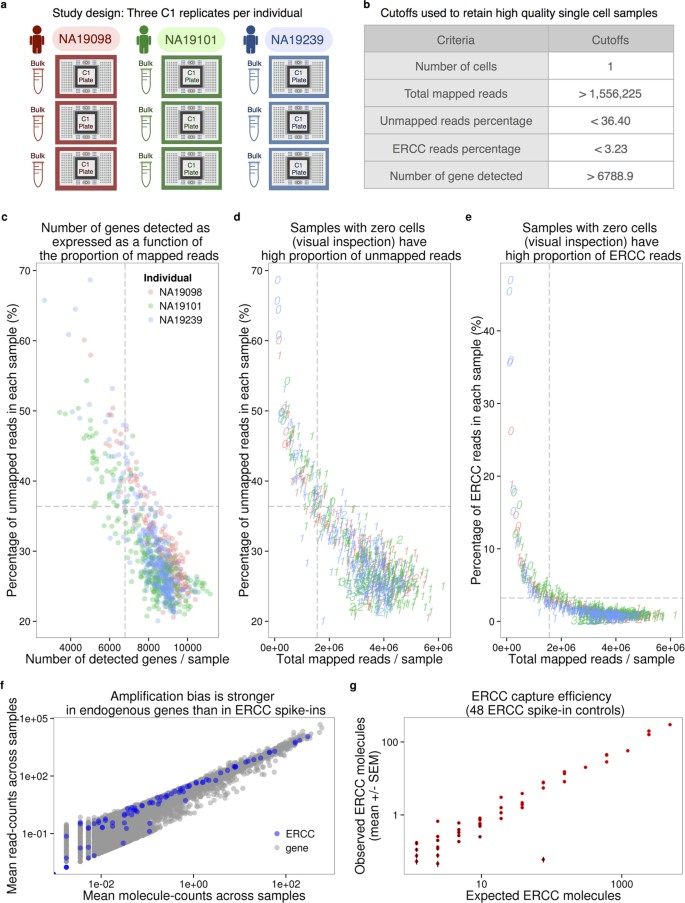
Highly parallel and efficient single cell mRNA sequencing with paired picoliter chambers | Nature Communications

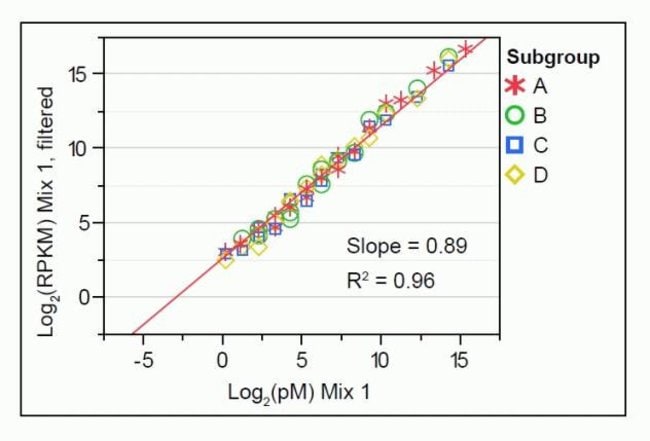
Science Mission - RNA capture sequencing of extracellular RNA from human biofluids Comprehensive transcriptome analysis of extracellular RNA (exRNA) purified from human biofluids is challenging because of the low RNA concentration and
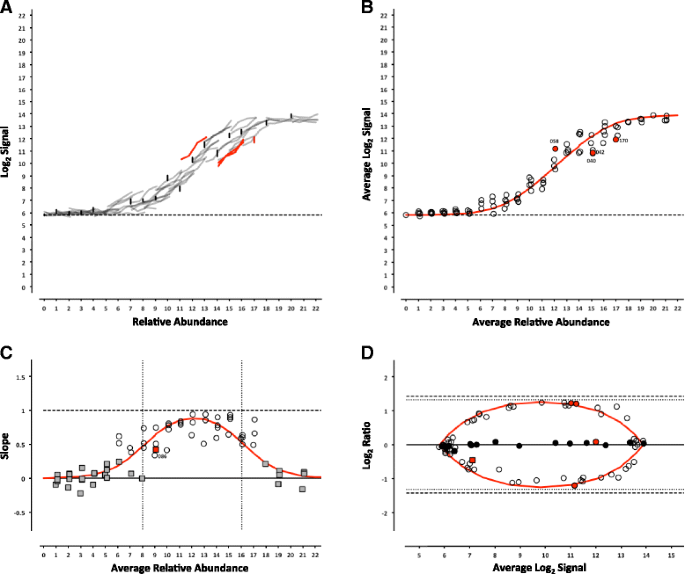
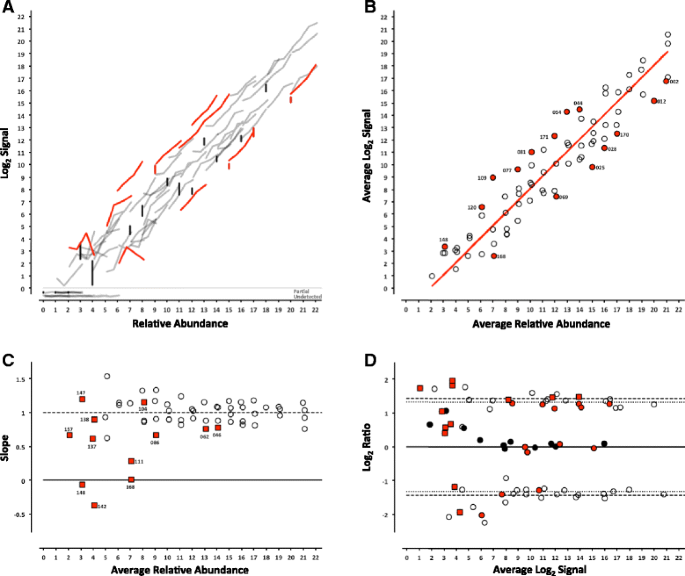
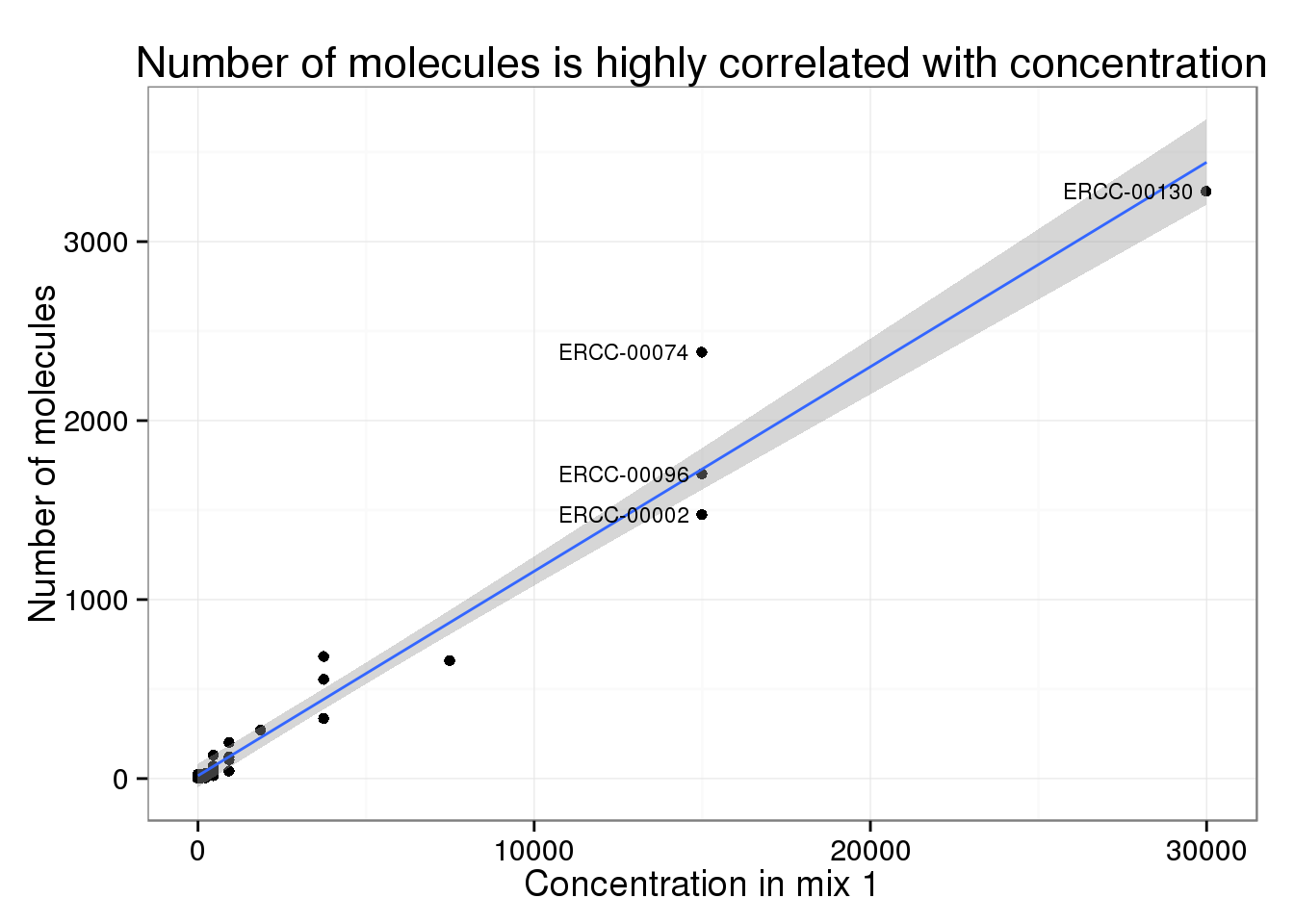
Evaluation of the External RNA Controls Consortium (ERCC) reference material using a modified Latin square design | BMC Biotechnology | Full Text
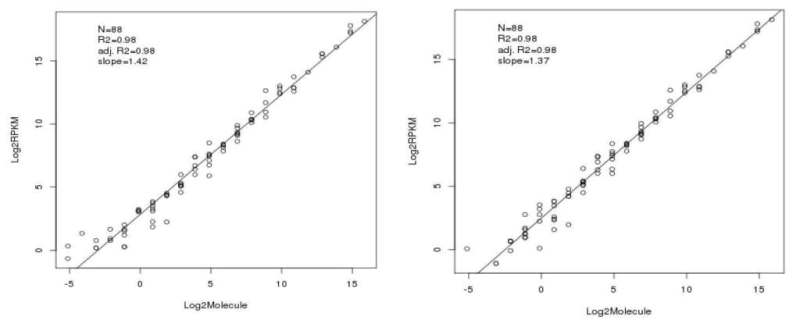
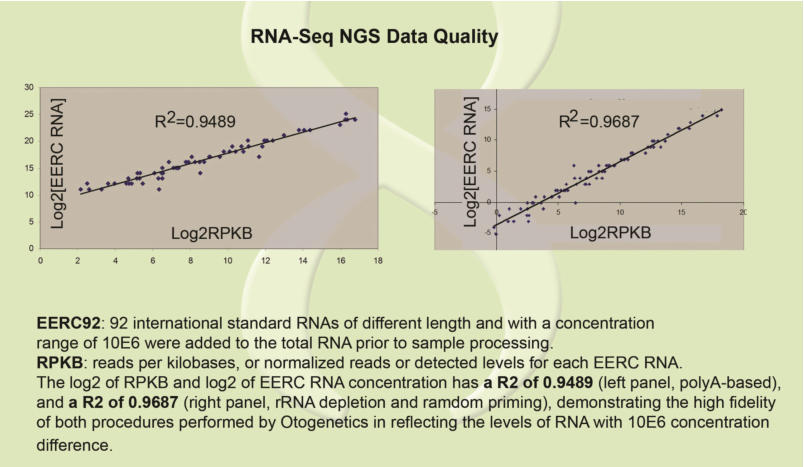
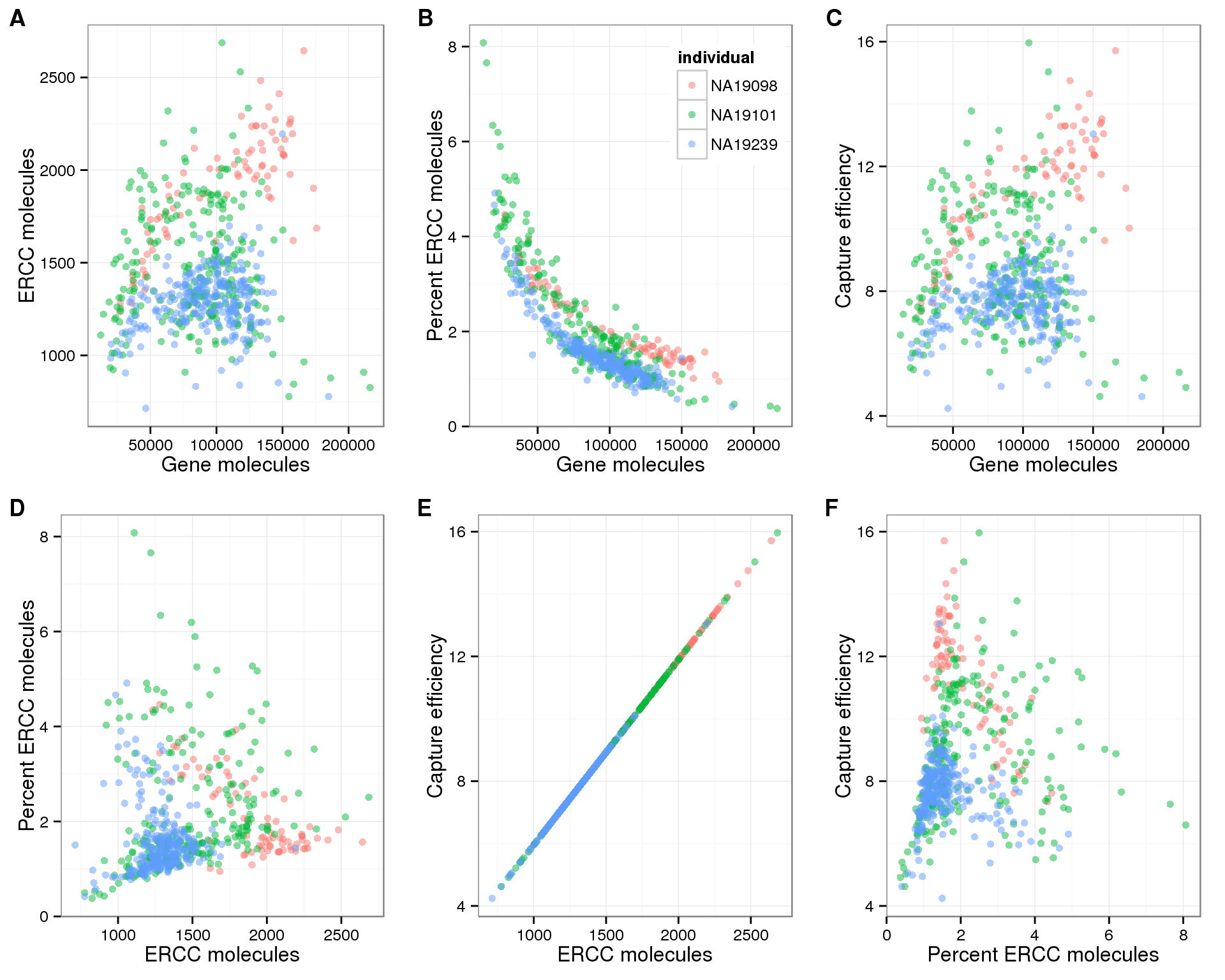
Using ERCC Spike-In Control Transcripts Provides Confidence in Agilent Microarray and RNA-Seq Gene Expression Data

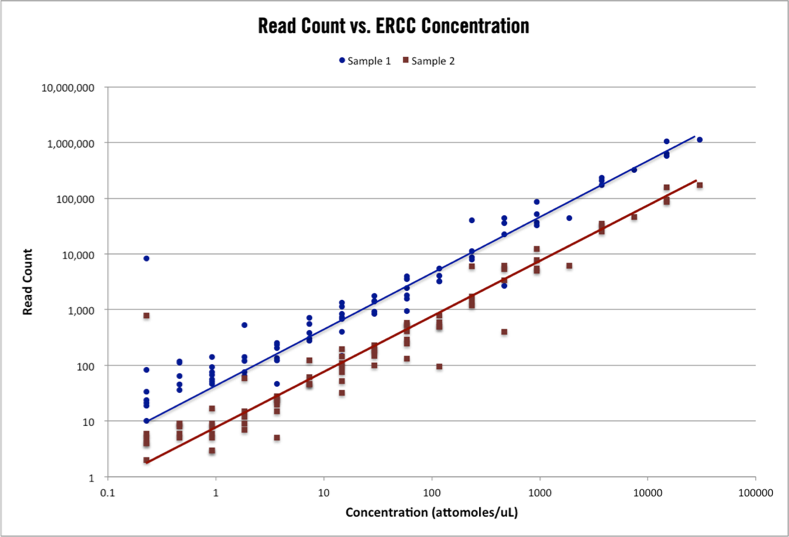
Benchmarking of the Oxford Nanopore MinION sequencing for quantitative and qualitative assessment of cDNA populations | RNA-Seq Blog

Assessing technical performance in differential gene expression experiments with external spike-in RNA control ratio mixtures | Nature Communications