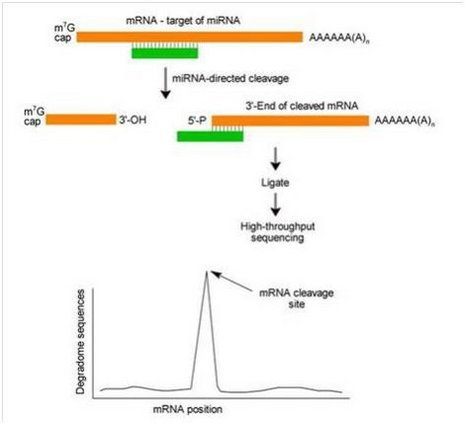
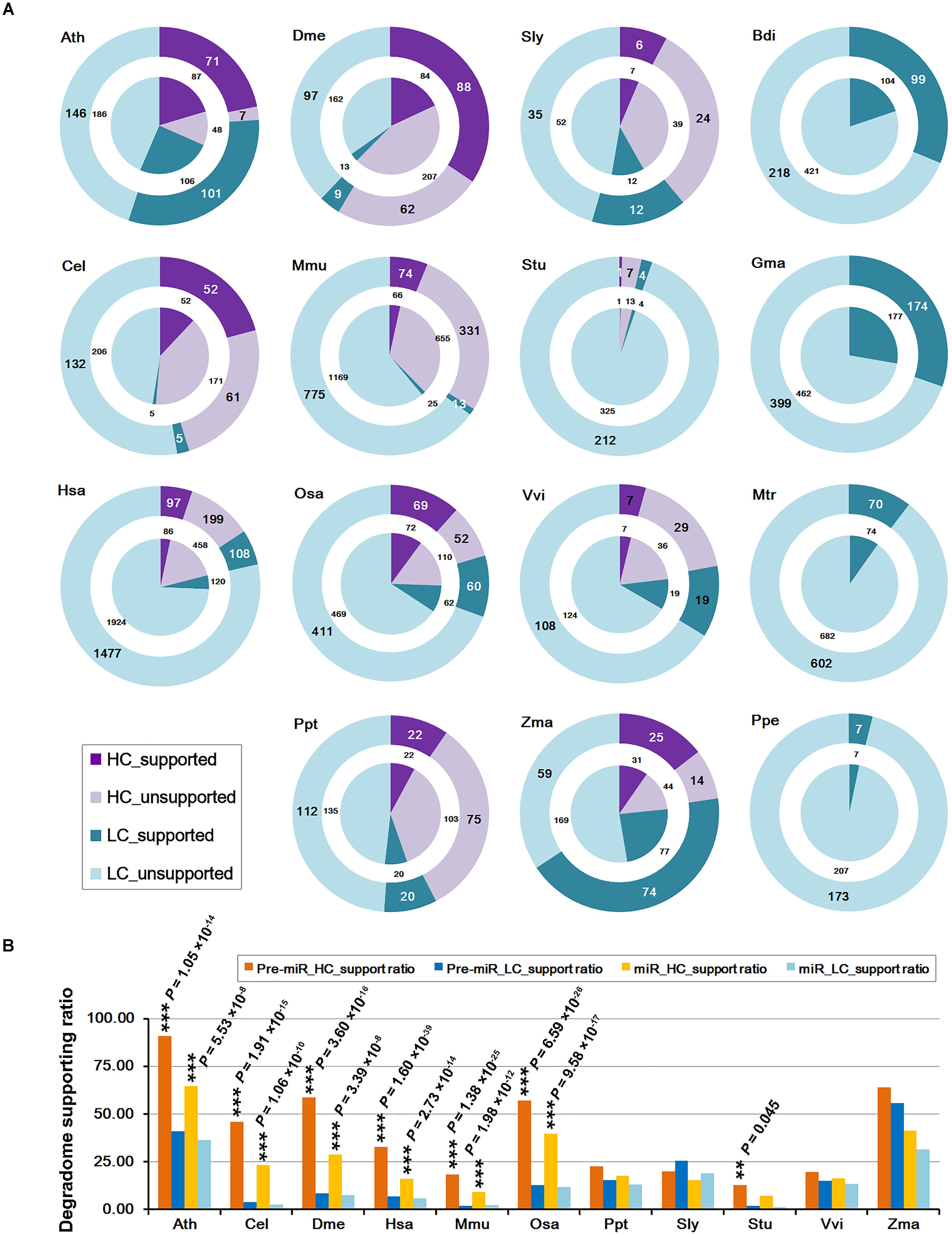
starBase: a database for exploring microRNA–mRNA interaction maps from Argonaute CLIP-Seq and Degradome-Seq data
Fast and Efficient 5'P Degradome Library Preparation for Analysis of Co-Translational Decay in Arabidopsis

High-throughput 5′P sequencing enables the study of degradation-associated ribosome stalls - ScienceDirect

Transcriptional multiomics reveals the mechanism of seed deterioration in Nicotiana tabacum L. and Oryza sativa L. - ScienceDirect

Comparative Small RNA and Degradome Sequencing Provides Insights into Antagonistic Interactions in the Biocontrol Fungus Clonostachys rosea | Applied and Environmental Microbiology
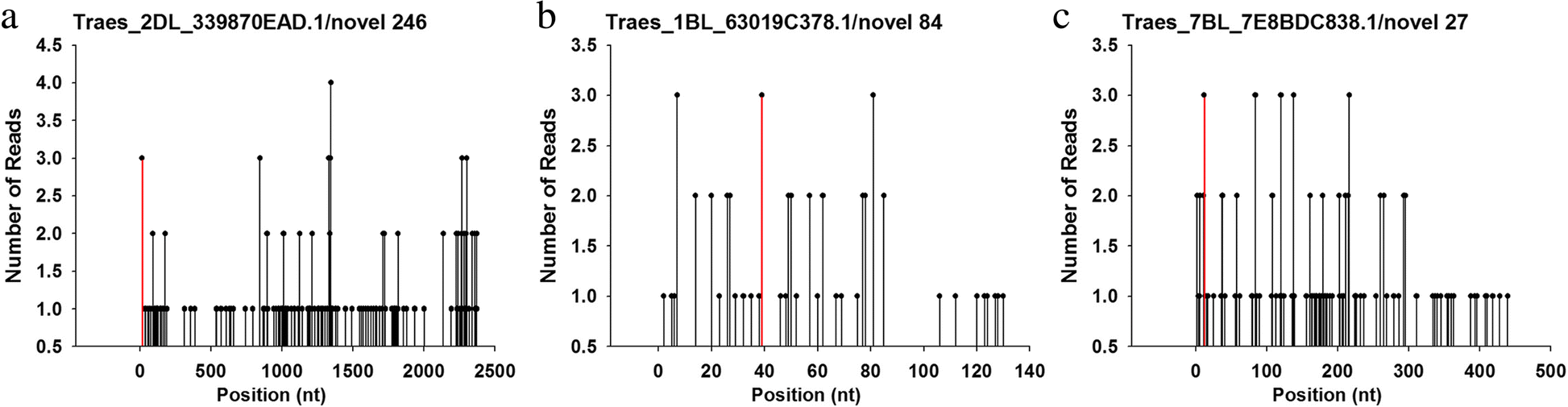
Small RNA and degradome sequencing used to elucidate the basis of tolerance to salinity and alkalinity in wheat | BMC Plant Biology | Full Text
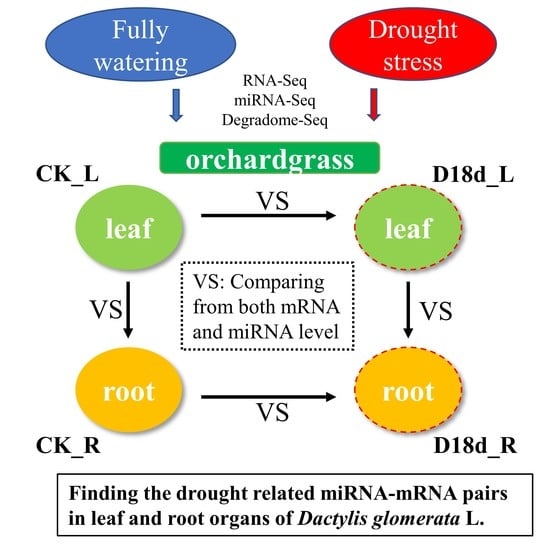
IJMS | Free Full-Text | Combinations of Small RNA, RNA, and Degradome Sequencing Uncovers the Expression Pattern of microRNA–mRNA Pairs Adapting to Drought Stress in Leaf and Root of Dactylis glomerata L.
Small RNA and Degradome Sequencing Reveal Complex Roles of miRNAs and Their Targets in Developing Wheat Grains | PLOS ONE

Integrated transcriptome, small RNA and degradome sequencing approaches provide insights into Ascochyta blight resistance in chickpea - Garg - 2019 - Plant Biotechnology Journal - Wiley Online Library

NMD-degradome sequencing reveals ribosome-bound intermediates with 3′-end non-templated nucleotides | Nature Structural & Molecular Biology

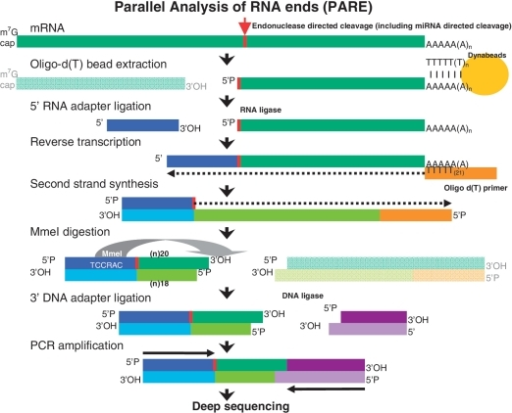
Construction of Parallel Analysis of RNA Ends (PARE) libraries for the study of cleaved miRNA targets and the RNA degradome | Nature Protocols

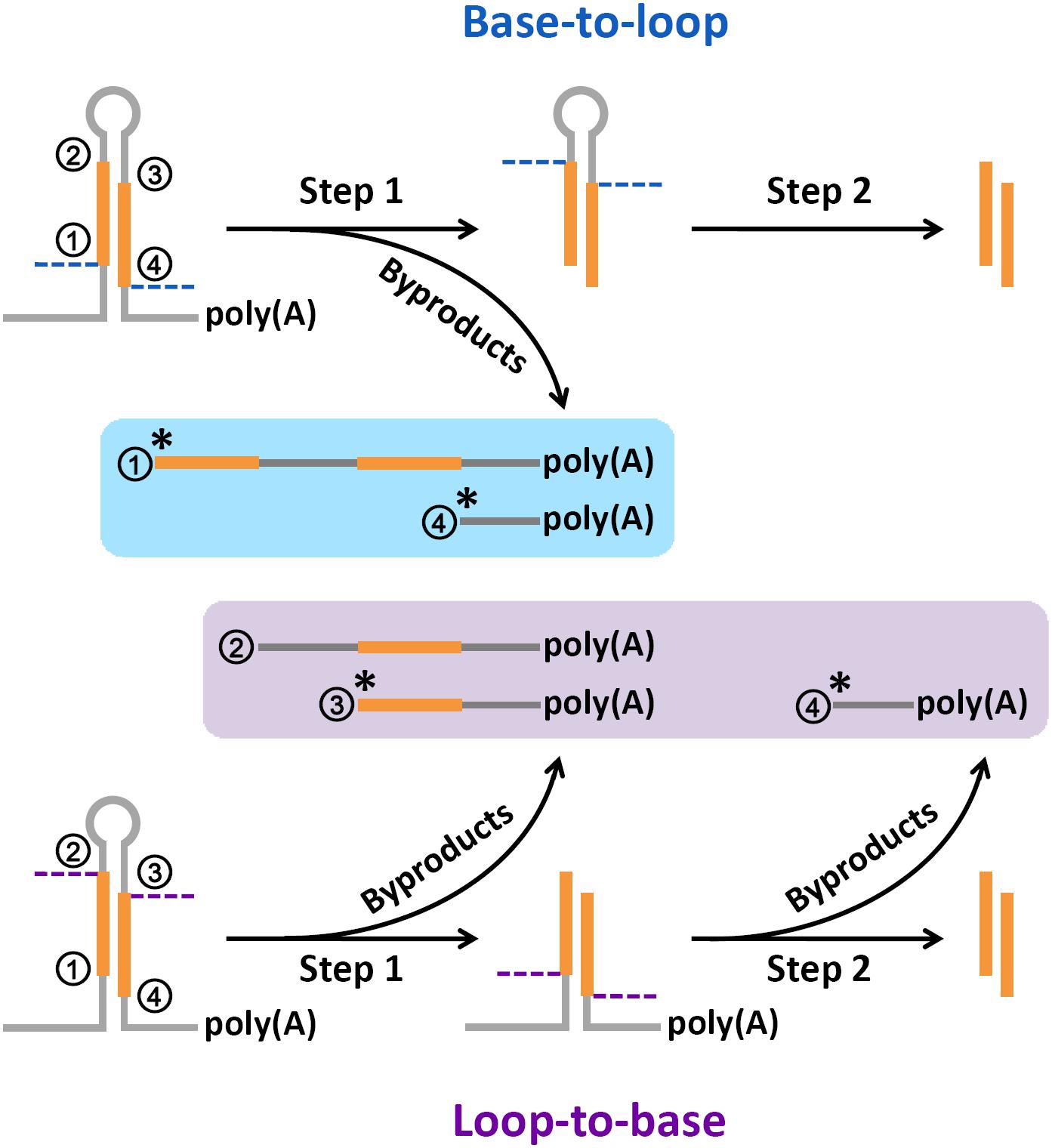
An improved method of constructing degradome library suitable for sequencing using Illumina platform | Plant Methods | Full Text

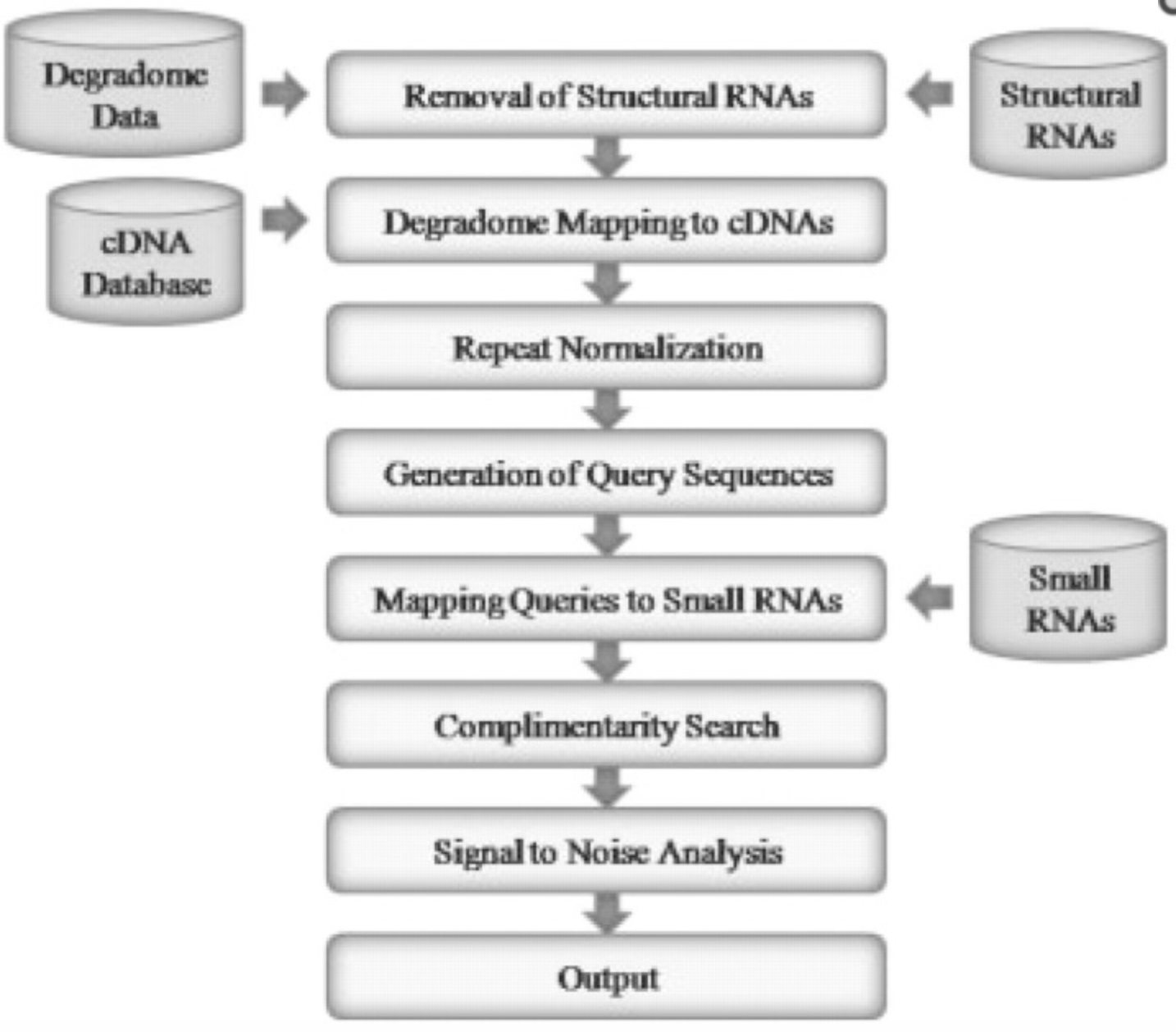
StarScan – a web server for scanning small RNA targets from degradome sequencing data | Web server, Data, Map reading