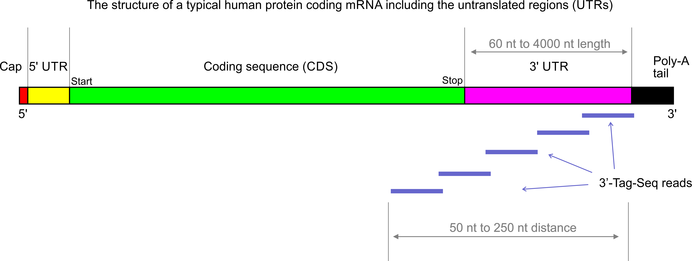
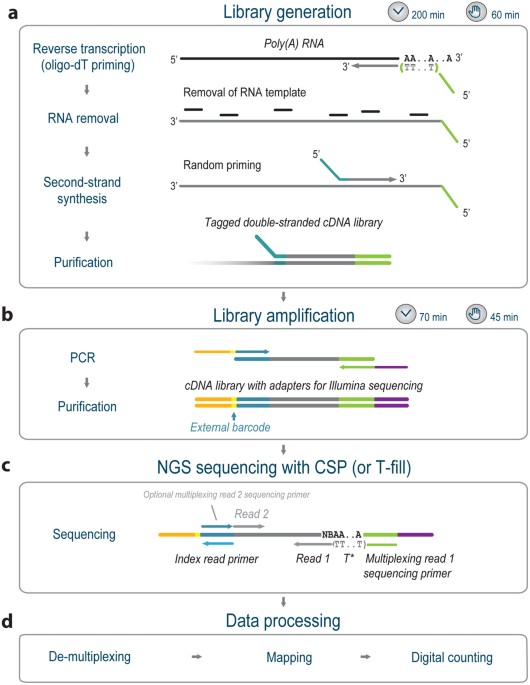
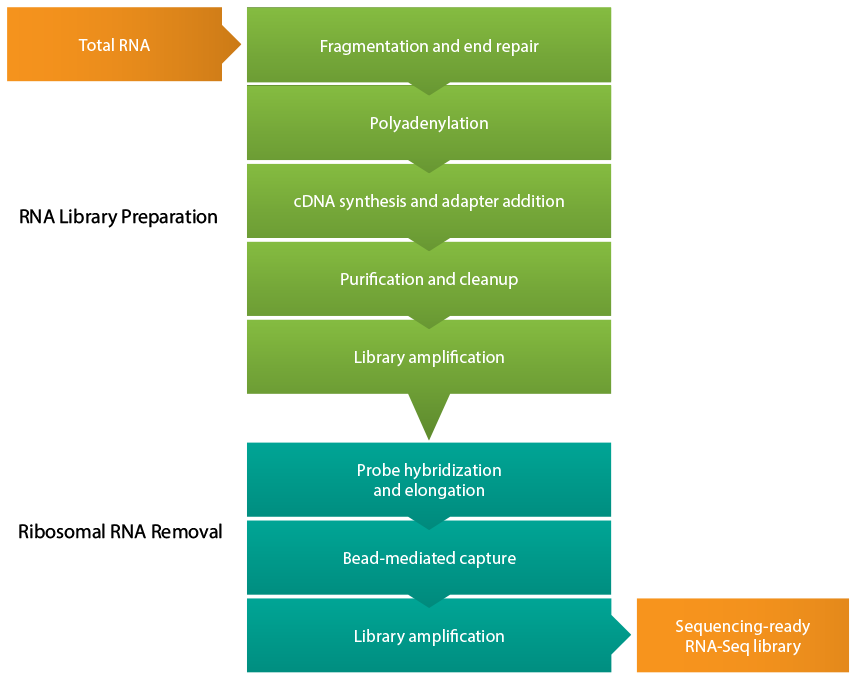
Figure 1 from A multiplex RNA-seq strategy to profile poly(A+) RNA: application to analysis of transcription response and 3' end formation. | Semantic Scholar
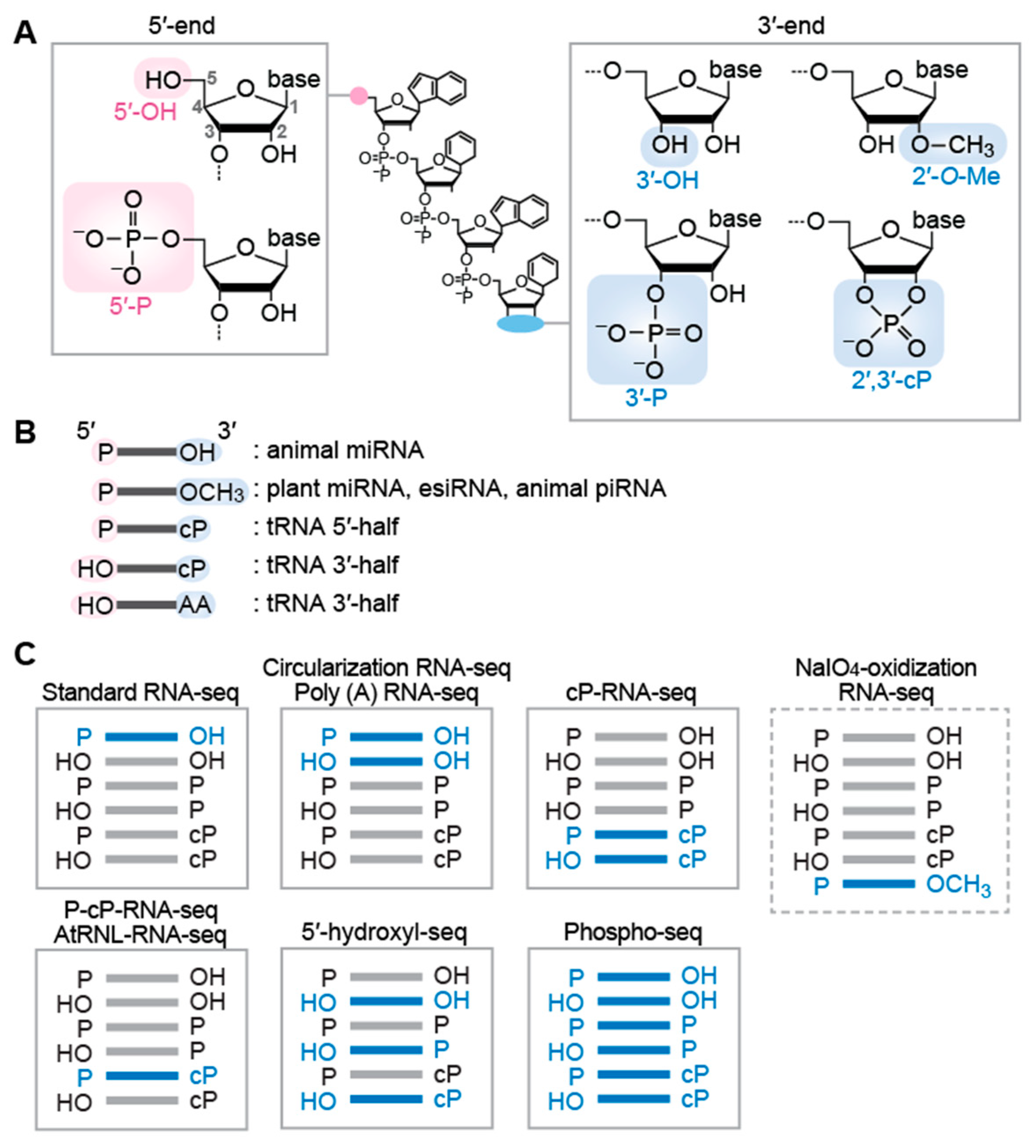
Biomolecules | Free Full-Text | Making Invisible RNA Visible: Discriminative Sequencing Methods for RNA Molecules with Specific Terminal Formations

The 'TranSeq' 3′‐end sequencing method for high‐throughput transcriptomics and gene space refinement in plant genomes - Tzfadia - 2018 - The Plant Journal - Wiley Online Library

Simultaneous measurement of transcriptional and post-transcriptional parameters by 3′ end RNA-Seq | RNA-Seq Blog

TM3'seq: a tagmentation-mediated 3' sequencing approach for improving scalability of RNA-seq experiments | bioRxiv
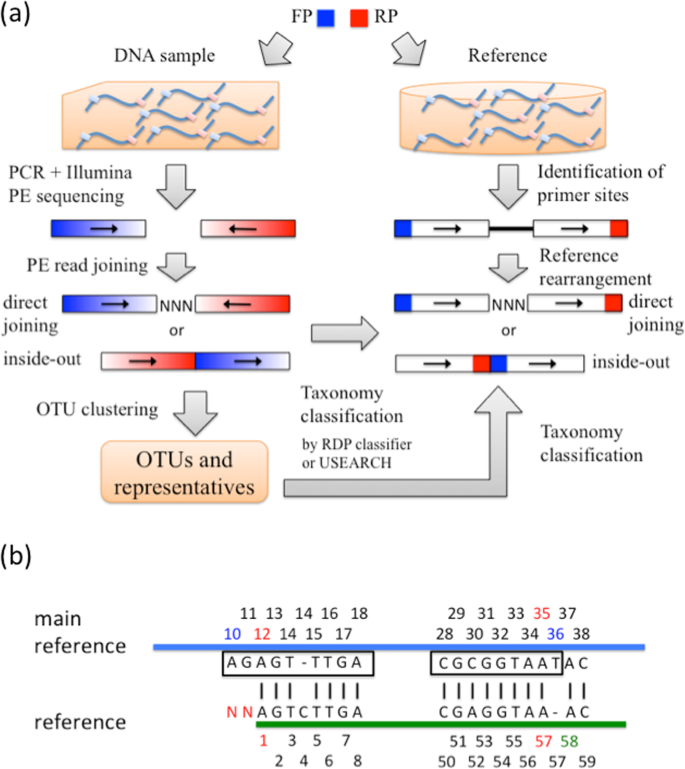
Joining Illumina paired-end reads for classifying phylogenetic marker sequences | BMC Bioinformatics | Full Text
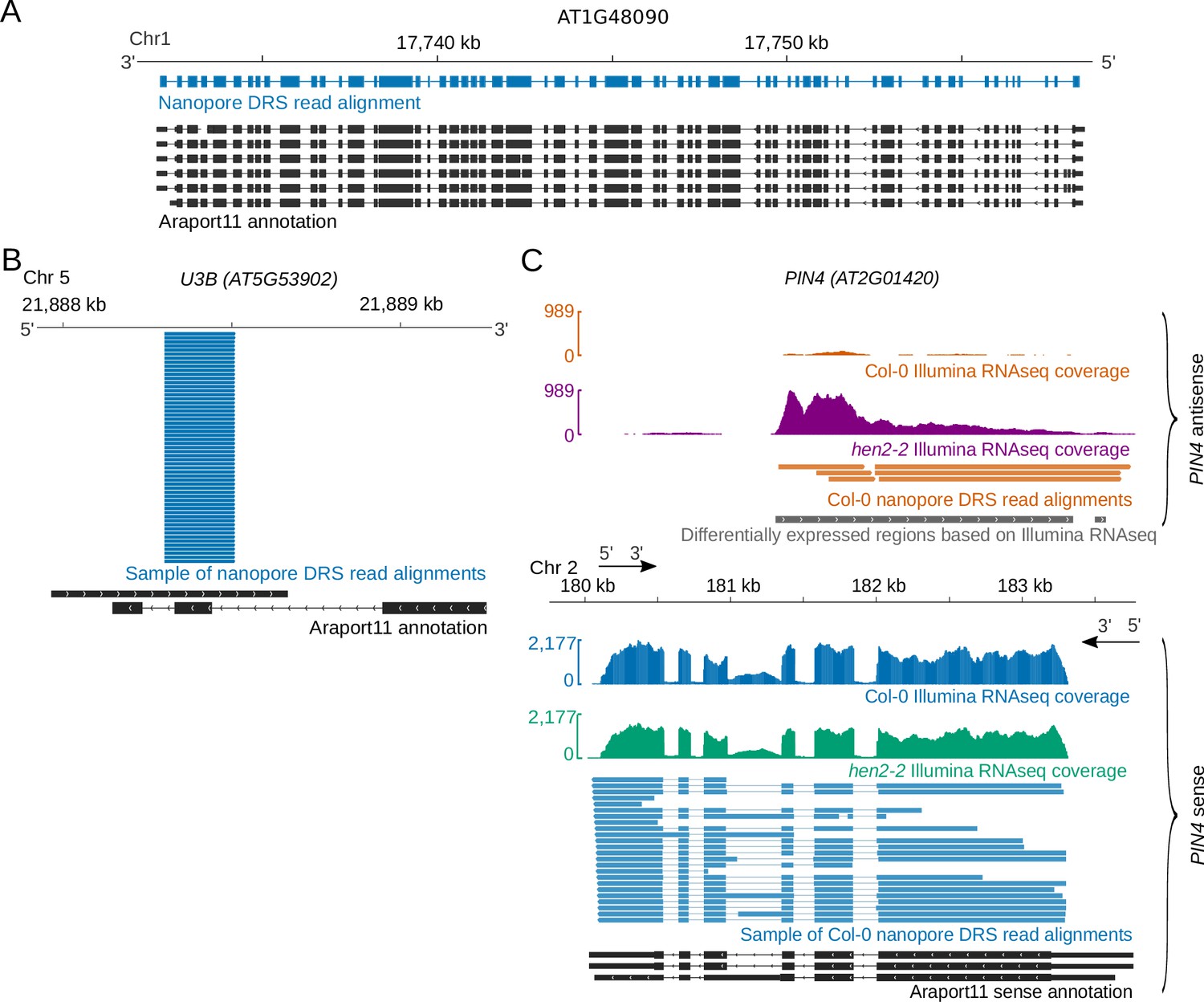
Nanopore direct RNA sequencing maps the complexity of Arabidopsis mRNA processing and m6A modification | eLife

Poly(A)-ClickSeq – click-chemistry for next-generation 3΄-end sequencing without RNA enrichment or fragmentation | RNA-Seq Blog

TRENDseq—A highly multiplexed high throughput RNA 3′ end sequencing for mapping alternative polyadenylation - ScienceDirect

3′ end sequencing workflow with oligo(dT) 19 V. a) Overview of the 3′... | Download Scientific Diagram

Nano3P-seq: transcriptome-wide analysis of gene expression and tail dynamics using end-capture nanopore cDNA sequencing | Nature Methods

General outline of oligo(dT)-based 3′-end sequencing protocols (e.g.,... | Download Scientific Diagram


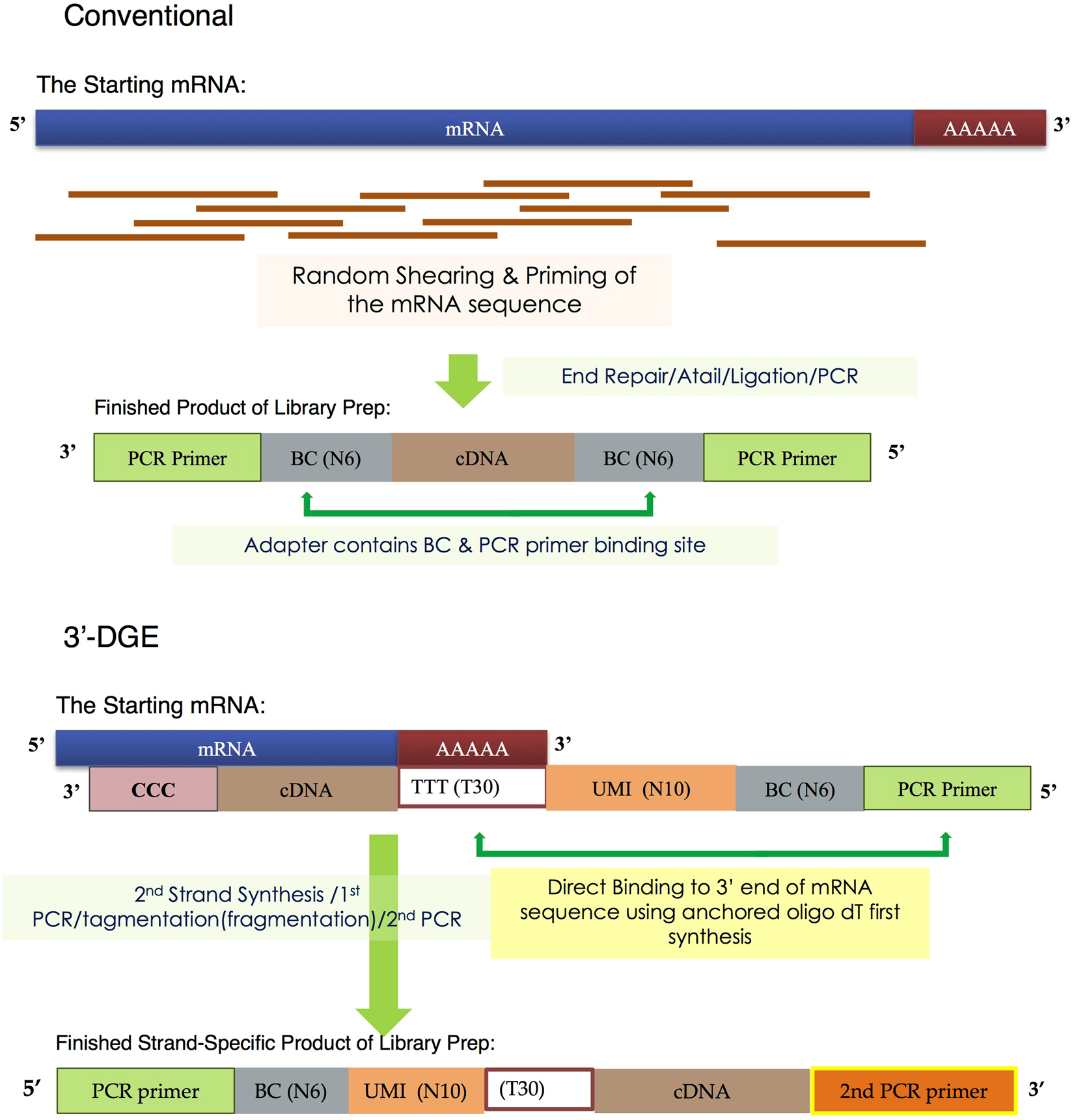
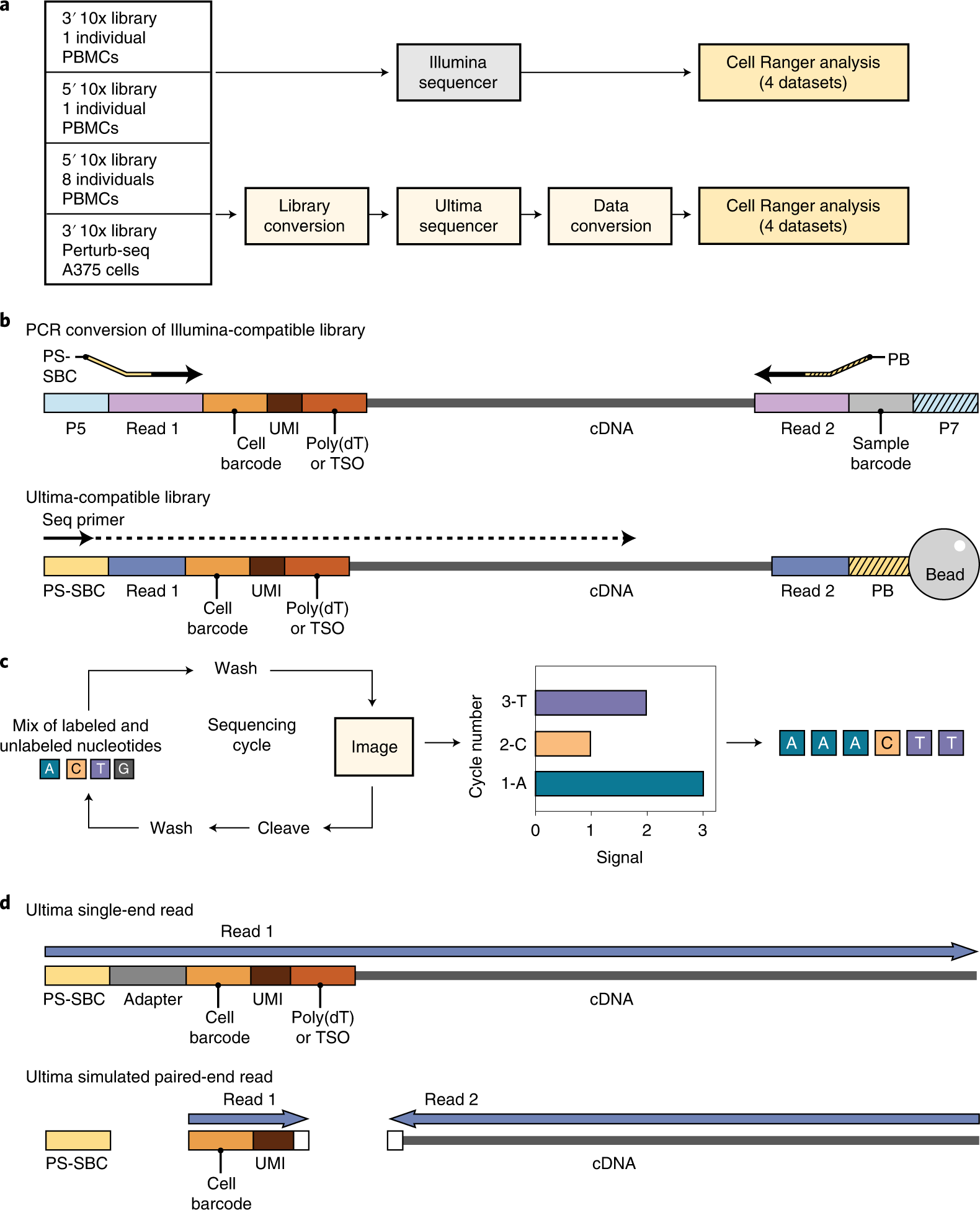
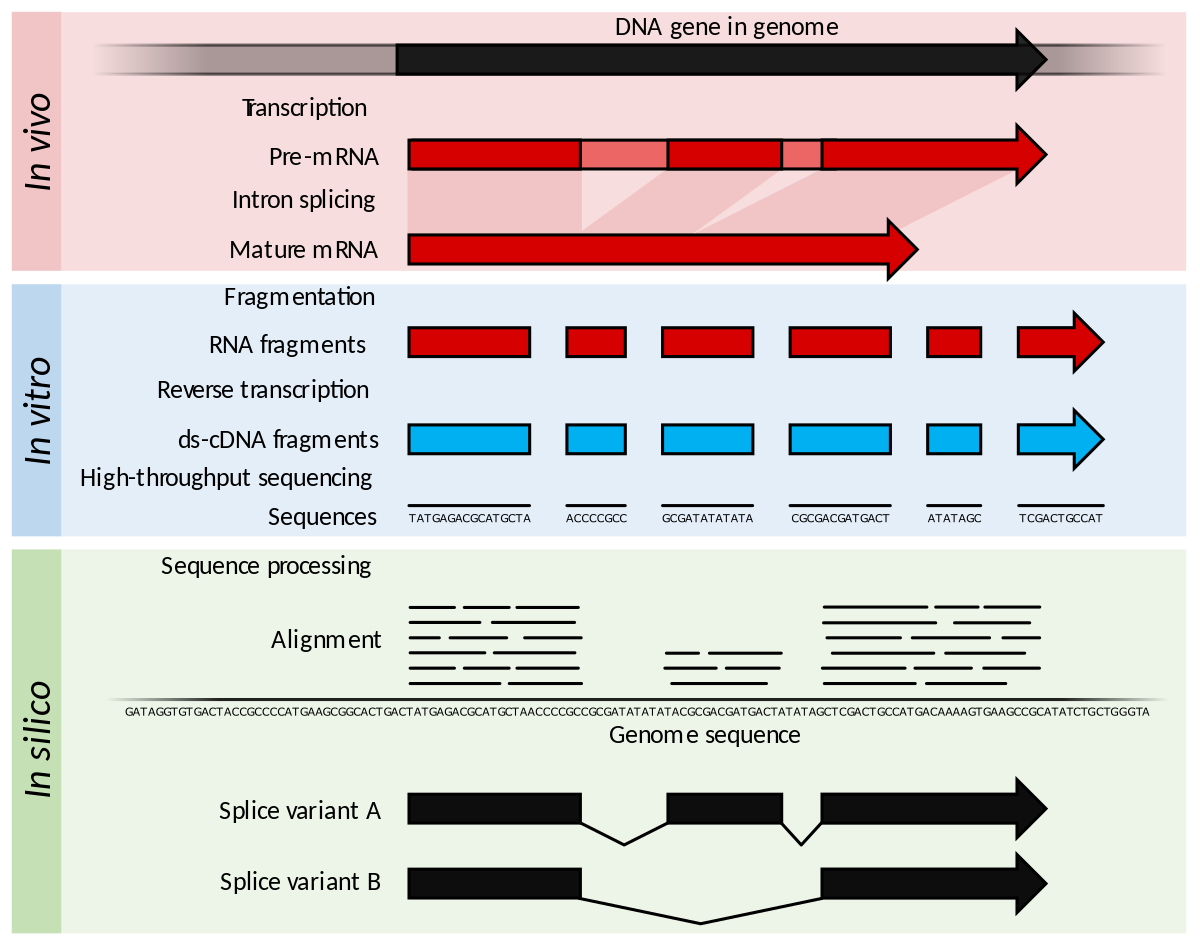

![3′ end sequencing methods. a In DeepSAGe [50] poly(A) + RNAs are... | Download Scientific Diagram 3′ end sequencing methods. a In DeepSAGe [50] poly(A) + RNAs are... | Download Scientific Diagram](https://www.researchgate.net/publication/262339133/figure/fig4/AS:667031334572034@1536044071441/end-sequencing-methods-a-In-DeepSAGe-50-polyA-RNAs-are-captured-by-oligo-dT.png)